Research Article Open Access
Tumor Genomic Profiling Reports from Different Vendors: A Comparison with Respect to Clinical Action Ability of the Provided Data
Yuriko Mori*, Victor Levenson and Jeffrey Otto
CHI Institute for Research and Innovation, Center for Translational Research, Baltimore, USA
- *Corresponding Author:
- Yuriko Mori
CHI Institute for Research and Innovation
Center for Translational Research
Baltimore, USA
Tel: 443-948-5435
Fax: 443-948-5421
Email: YurikoMori@gmail.com
Received date: July 03, 2016; Accepted date: August 02, 2016; Published date: August 05, 2016
Citation: Mori Y, Levenson V, Otto J (2016) Tumor Genomic Profiling Reports from Different Vendors: A Comparison with Respect to Clinical Action Ability of the Provided Data . Adv Mol Diag 1:110.
Copyright: © 2016 Mori Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Advances in Molecular Diagnostics
Abstract
Background: Next generation sequencing (NGS) of selected genes is an expanding field of solid tumor characterization. Multiple vendors offer this service, but panel design and policies on interpretive reporting are variable. This study compared reports from selected vendors, with emphasis on clinical Action ability reporting.
Methodology: DNA aliquots of five breast and five colorectal cancers were sent to providers offering the following solid tumor NGS tests: Foundation oneTM, StrandAdvantageTM, CANCP, and GeneTrailsTM. The interpretive reports were compared for the reporting of clinically actionable variants.
Results: Detection of mutants was mostly consistent (17 of 21, 81% concordant), while detection of copy number variants was highly discordant (6 of 23, 26.1% concordant). Discordance of copy number variation appeared to be primarily due to difference in analytical sensitivity. Actionable variants with approved targeted therapies for the same tumor type were detected and interpreted uniformly by all tests. Reporting of variants with other action ability was variable, largely due to difference in panel design and analytical sensitivity, and, to a lesser degree, to different annotation policies. Actionable variant reporting by Foundation one provided better coverage of clinical trials for targeted therapies while strand advantage uniquely covered chemotherapeutic response modulation. GeneTrails provided moderate coverage of clinical trials despite its small gene panel size. Utility of rearrangement targets in Foundation one and StrandAdvantage panel was not evident, due to the tumor type assessed.
Conclusion: All enrolled tests reported actionable variants with FDA-approved target therapy for the specific tumor type with consistent clinical interpretations. In contrast, reporting of variants with other Action ability was heavily impacted by panel design, reporting policy, and annotation policy. Larger studies will be needed to quantify the qualitative differences outlined above and evaluate the benefit to the patient in order to improve the practice of precision medicine in the future.
Introduction
Genetic profiling of cancer aims to identify molecular variants in the tumor that might help guide patient management. For instance, it is recommended that KRAS and NRAS genes be genotyped in metastatic colon cancer tissue and patients with mutations in codons 12, 13 of exon 2, codons 59, 61 of exon 3, and codons 117 and 146 of exon 4 should not be treated with either cetuximab or panitumumab [1-3]. Similarly, amplification of the MET gene is also associated with poor response to cetuximab and panitumumab therapy [4-10]. Large clinical studies have found that a mutation in codon 600 of BRAF is a negative prognostic factor in metastatic colorectal cancer [11,12].
Increasing utility of multi-gene tumor profiling for patient management has led to an increase in the utilization of Next- Generation Sequencing (NGS) assays in order to identify clinically informative variants in multiple genes. These complex assays require careful planning on multiple design parameters: panel design (i.e. selection of genes and number of targets to include), assay type, NGS platforms, variant calling algorithms, thresholds to call Copy Number Variation (CNV) and rearrangements. Additionally, multiple interpretive considerations need to be included in this planning (e.g. how to interpret the oncogenicity of a variant, or its ability to activate a particular pathway, and what clinical utility to report for the variant based on literature assessment).
Different vendors make different choices along each of these dimensions. For instance, some vendors use small sequencing panels of up to approximately 50 genes, often choosing to sequence only mutation hotspot regions in these genes. Yet others use larger panels of hundreds of genes, choosing to sequence whole genes. Some vendors use the Illumina platform, others the Life Technologies platform, and yet others-the Roche 454 platform. Selection of algorithms for calling variants, amplifications, deletions and translocations to a large degree depends on the workflow upstream of sequencing and is different for amplicon sequencing and hybridization-based capture methods. The next step is the rather difficult task of curating literature and various databases to determine if a variant is indeed oncogenic, and if so, whether studies indicate that it modulates response to any of the several applicable therapies in the tumor type of interest. There are several choices in this process as well: what threshold of study size is deemed to provide sufficient evidence of clinical utility, whether to include studies on other cancer types as the supporting evidence, whether to use an inclusive or a restrictive approach in interpreting the clinical utility of variants whose significance is not fully clear, how to handle “off-label” use of Food and Drug Administration (FDA)- approved drugs, etc.
Given this plethora of choices, it is important to understand how these choices reflect in the final report that an oncologist receives for the case at hand. Do the clinical recommendations in these reports vary widely based on these choices or are they largely consistent irrespective of these choices? To the best of our knowledge, there hasn’t been a formal study addressing this question. Here we present an analysis of clinical reports for 10 cases, sent to 4 different vendors, who have made different choices in designing their cancer profiling assays and the subsequent interpretation and reporting protocols.
Materials and Methods
Clinical samples and DNA
De-identified formalin-fixed, paraffin-embedded (FFPE) tumor tissues from 5 CRC (Colorectal Carcinoma) and 5 hormone receptor (HR)-negative breast cancer (BC) from the databank at the CHICenter for translational research were chosen for the study. All but one BC samples was triple negative breast cancer (TNBC). All specimens demonstrated tumor cell percentages greater than 50%. This study was conducted under a protocol approved by Western Institutional Review Board (Puyallup, WA). Written consent was obtained from all patients. DNA was extracted from the samples at the CTR Molecular Diagnostic Laboratory using the QiaAmp FFPE DNA isolation kit (Qiagen, Germany) without macrodissection. The harvested DNA was assessed for quality by using NanoDrop 2000 spectrophotometer (Thermo Scientific; Wilmington, DE) and for double stranded-stranded DNA concentration by using Qubit 2.0 (Thermo Scientific), and Qubit dsDNA HS assay kit (ibid) in order to verify that DNAs met. The quality standard that were requested by each vendor. Once measurements were completed, DNAs were aliquoted and stored at -20°C until shipment.
NGS assays
The following four tests were used:
The Foundation one gene panel from Foundation Medicine Inc. (Cambridge, MA) comprises 315 genes as well as introns of 28 genes involved in rearrangements. This is a hybridization-based capture panel, with sequencing performed on an Illumina platform [13].
The CANCP panel from the Mayo Medical Laboratories at Mayo Clinic (Rochester, MN) has 50 genes. It is a PCR-based amplicon hotspot panel, with sequencing performed on the Life Technologies Ion PGM platform. Unlike other tests, CANCP doesn’t assess Copy Number Variation (CNV).
The GeneTrails solid tumor panel from the Knight Diagnostic Laboratory, at the Oregon Health and Science University (Portland, OR) has 37 genes. This is a PCR-based amplicon sequencing panel, sequenced on the Life Technologies Ion PGM platform.
The StrandAdvantage panel from Strand Genomics Inc (Aurora, CO) comprises 151 genes and rearrangements in the ALK, ROS and RET genes. This is also a hybridization-based capture panel, with sequencing performed on an Illumina platform.
Targets assessed by each test are listed in supplementary Table 1. A frozen DNA aliquot per sample was shipped out to each sequencing vendor, with de-identified clinicapathological data, according to each vendor’s specification. Although FFPE tissue was the preferred submission format for all vendors, DNA submission was selected for this study in order to eliminate data variability arising from FFPE section unevenness. Additionally, a representative HE-stained tissue section for each block was provided to each vendor, each vendor performed its own quality control of the received aliquots prior to NGS assay. The NGS assay and reporting was conducted from December, 2014 to April, 2015. Thus, NGS assay and reporting described in this study reflect each vendor’s practice during this period. The list of targets assessed by each test is available in supplementary Table 1. Specific genetic regions assessed were unavailable and were not considered in our analyses. Interpretive NGS reports received from these vendors including the limit of detection data, when permission for publication is granted by the vendor, are available as part of supplementary material S1.
| Sample ID | Cancer Type | Sex | Age | Stage | Foundation one | StrandAdvantage | GeneTrails | CANCP |
|---|---|---|---|---|---|---|---|---|
| 202 | CRC | F | 80 | IIIC | Y | Y | Y | Y |
| 240 | CRC | F | 82 | IIC | Y | Y | Y | Y |
| 289 | CRC | M | 78 | IIIC | Y | Y | Y | Y |
| 80 | CRC | F | 92 | IIA | Y | Y | Y | Y |
| 38 | CRC | F | 84 | IIIB | Y$$ | Y | N | N |
| 241 | TNBC | F | 74 | IIB | Y | Y | Y | Y |
| 275 | TNBC | F | 56 | IIA | Y | Y | Y | Y |
| 84 | TNBC | F | 50 | IIA | Y | N | Y | Y |
| 358 | TNBC | F | 29 | IIA | Y$ | Y | N | Y |
| 26 | HR-/HER2+ BC | F | 79 | IIIC | Y | N | Y | Y |
Table 1: Samples used in this study and their report availability.
Analysis of reports
These reports were compared across the four tests, with primary focus on clinically actionable variants. For the purposes of comparison, such variants were defined as those that fall in one of the following 4 categories.
Variants modulating targeted therapy approved for use in the same tumor type
Variants modulating targeted therapy approved for use in another tumor type
Variants modulating targeted therapy currently in trials
Variants modulating efficacy or toxicity of chemotherapy and/or radiotherapy
Reported variants were excluded from our analysis if all reporting vendors designated them as: a) variants of unknown significance (VUS), or b) variants with clinical significance that didn’t confer any of the four forms of actionablity.
The variants and their action ability annotations were then compared, with follow up literature review wherever required to assess the accuracy of the Action ability ascribed. For test reports that provided summary and detailed narrative separately (Foundation one and Strand Advantage), annotations given in the summary were used for the analysis. Discrepancies between summary and narrative were noted if existed. For test reports that did not always provide explicit statement on FDA-approved tumor type (GeneTrails and CANCP), we assigned the category according to the current approval status in order to enable the comparison. Erroneous FDA-approval status assignment in reports was also corrected based on our literature review for the purpose of comparison.
Results
Characteristics of reports
Not all of the 10 chosen samples ran successfully on all tests; 1-2 samples for each vendor were dropped due to the factors beyond the vendors’ control, such as sample exhaustion or compromised sample during shipment. Table 1 shows sample description and report availability.
Foundation one provided reports for all 10 samples, but inspection of initial reports revealed that samples #358 (TNBC) and #38 (CRC) were interchanged by the vendor at a pre analytical stage. Reanalysis and acquisition of corrected report were done for sample #358 but not for sample #38 due to sample exhaustion. Thus, the Foundation One report for sample #38 was dropped from the analysis. Of note, DNA submission was a non-preferred sample format for all vendors, thus pre analytical workflow in this study was not their standard practice.
All tests included actionable variants, as well as VUS and clinically significant variants without Action ability (defined in the Method section), in their reports. Some differences in reporting were observed; a) CANCP didn’t assess CNVs, b) Strand Advantage reported germline polymorphisms, based on the population allele frequency and variant allele prevalence in the tumor, if they were deemed actionable, c) Foundation One stated that the Action ability annotations were “not evaluated for source or level of published evidences,” d) GeneTrails and CANCP did not always explicitly state the FDA-approval status of the referred agents (i.e. approved for the patient’s or other tumor types), e) CANCP mentioned potential targeted therapies under research but did not indicate whether pertinent clinical trial existed or not, f) Foundation One and Strand Advantage provided detailed clinical trial information (e.g. trial ID, title, and phase), while Gene Trails provided only trial ID, g) Gene Trails did not provide supporting literature for clinical significance, and h) Foundation One and Strand Advantage provided summary of detected variants and their action ability separately from narrative interpretation. Median report length in page number greatly differed among vendors; 16 for Foundation one, 9.5 for StrandAdvantage, three for GeneTrail, and four for CANCP. The difference in report length was attributed to factors e)- h) listed above. Table 2 lists the clinically actionable variants reported for each sample by each vendor, along with their Action ability annotation according to the four criteria defined in the Method section.
| Sample ID | Type | Foundation one | StrandAdvantage | GeneTrails | CANCP |
|---|---|---|---|---|---|
| 202 | CRC | MET amplification-equivocal (II,III), TP53 p.Arg196X (III) | MET amplification (II,III), ERCC2 p.Lys751Gln (IV), ERCC5 p.Asp1104His (IV), XRCC1 p.Gln399Arg (IV) | TP53 p.Arg196X and deletion (III) | No actionable variants |
| 240 | CRC | SMO p.Ala601Val (II#,III#), TP53 p.Arg248Gln, p.Arg282Trp (III) | AURKA amplification (III), EGFRamplification (I), TP53 p.Arg248Gln, p.Arg282Trp (IV), APC p.Ala1366SerfsX8 (IV), XRCC1 p.Gln399Arg (IV) | TP53 p.Arg248Gln, p.Arg282Trp (III) | No actionable variants |
| 289 | CRC | BRAF p.Val600Glu (I,II,III), PTCH1 p.Val692fsX54 (II,III), FBXW7 p.Arg465His (III), PTEN rearrangement intron 1 (III), TP53 p.Gly244Cys (III) | BRAF p.Val600Glu (I,II,III),MLH1 c.-93G>A (IV) | BRAF p.Val600Glu (I/II,III), TP53 p.Gly244Cys and deletion (III), TSC2 deletion (I/II) and TP53 deletions | BRAF p.Val600Glu (I/II), |
| 80 | CRC | KRAS p.Gly13Asp (I,II,III), PIK3CA p.Asn345Ser (III#), PIK3R1 p.Lys567Glu (III) | KRAS p.Gly13Asp (I,II,III), APC p.Arg232X p.R1114X (IV), XRCC1 p.Gln399Arg (IV) | KRAS p.Gly13Asp (I/II,III), PIK3CA p.Asn345Ser(III), PIK3R1 p.Lys567Glu (III), TP53 deletion (III) | KRAS p.Gly13Asp (I/II) |
| 38 | CRC | No Report | MTOR p.Glu1799Lys p.Thr1977Lys (II), KRAS p.Ala146Thr (II,III), APC p.Thr1445GlnfsX28 p.Gln1469X p.Arg216X (IV) | No Report | No Report |
| 241 | TNBC | NOTCH2 p.Arg2400X (III), TP53 p.Trp53X (III) | JAK2 amplification (II,III), KRAS amplification (II,III) , MYC amplification (III), TP53 p.Trp53X (IV), ABCC1 c.825T>C (IV) | TP53 p.Trp53X (III) | No actionable variants |
| 275$ | TNBC | PIK3CA p.Asn1068fsX3+ (I#*,II#,III#), PTEN p.Gly132Asp (I#*,II#,III#), CDK6 amplification (III), TP53 p.Tyr163X (III) | PTEN p.Gly132Asp (II,III),TP53 p.Tyr163X (IV), ABCC1 c.825T>C (IV), DPYD p.Met166Val (IV) | PTEN p.Gly132Asp (I/III) | No actionable variants |
| 84 | TNBC | FGFR1 amplification (II,III), CCND2 amplification (III), CCNE1 amplification (III), CDKN2A/B loss (III), MYC amplification (III), MYCL1 amplification (III), TP53 p.Val122fsX26 (III) | No Report | FGFR1 amplification (III),CDKN2A/B loss (III), TP53 p.Val122fsX26 (III) | No actionable variants |
| 358 | TNBC | AKT3 amplification–equivocal (I*,II,III), TP53 p.Gln165X (III) | AKT3 (II,III), DDR2 (II,III), FGFR1 (III), FGFR2 (III), MET (II,III), SMO (II,III), NTRK1amplifications (III), BRCA2deletion (II,III,IV), TP53 p.Gln165X (IV), DPYD p.Met166Val (IV) | No Report | No actionable variants |
| 26 | HR-/HER2+BC | ERBB2 amplification (I,II,III), PIK3CA p.Cys420Arg (I*,II,III), AURKA amplification (III), BARD1 p.Lys209fsX4 (III), GNAS amplification (III), MYC amplification (III), TOP2A amplification (III) | No Report | ERBB2 amplification (I/II,III), PIK3CA p.Cys420Arg (III) | No actionable variants |
I: Targeted therapy approved in the same tumor type. II: Targeted therapy approved in another tumor type. III: Targeted therapy in trials. IV: Chemotherapy/Radiotherapy efficacy or toxicity. I/II: Targeted therapy approved, but approved tumor type not provided in the report (Bold font denotes the category base on our literature review). # The Action ability annotations in summary and narrative are discrepant. Narrative refers to these variant as “the mutation reported has not been characterized.” Inadequate category label ofEverolimus (category I for TNBC), which is approved only for hormone receptor positive, HER2 negative breast cancer (category II).$ GeneTrails reported TP53 p.Y153X in #275 as non-actionable clinically significant variant, despite it reports TP53 loss of functionmutations in other TNBCs as actionable.
Table 2: Variants reported as clinically actionable in each report.
This table doesn’t include variants reported as VUS or as nonactionable.
Detailed review revealed some apparent errors in reporting, (see Table 2 for specific events). Discrepancy was noted in several annotations of clinical significance in the summary table and narrative of Foundation one reports (i.e. “actionable” in summary and “mutation reported here has not been characterised,” which is indicative of VUS, in narrative; for SMO p.Ala601Val, PIK3CA p.Asn345Ser, PIK3CA p.Asn1068fsX3+, and PTEN p.Gly132Asp). This finding might imply that Foundation one may not always exclude functionally illcharacterized variants prior to consider clinical action ability. Foundation one also mislabelled the FDA-approval status of Everolimus as approved in TNBC in two instances (Action ability category I), despite it being approved only for HR+ HER2- breast cancers (Action ability category II). (http://www.accessdata.fda.gov/drugsatfda_docs/nda/2012/022334Orig1s016.pdf). GeneTrails had discrepant action ability annotation of loss of function mutations in TP53 for stage IIA TNBCs (i.e. category III-actionable in sample #84 and #241, versus non-actionable in sample #275).
For the subsequent analyses, we corrected FDA-approval status error/ambiguity in the reports to the actual approval status. Intravendor annotation discrepancies were left unrevised, since it is impossible to surmise which of the conflicting statements represents the vendor’s standing at the time.
Test-to-test comparison in variant detection
Sixty variants were reported as clinically actionable by at least one test in this study. Thirty-nine of these variants were assessed, at least in one sample, by multiple tests that were capable of reporting them based on the panel design and vendor’s reporting policy. A total of 44 variant detection events met this description, and could be evaluated for test-to-test consistency in variant detection. Discordance among tests was significantly more frequent for detection of CNV (17 of 23, 73.9%) than mutations (4 of 21, 19.0%; p<0.01, Fisher’s exact test). There was no significant difference in discrepancy rate based on the type of mutation (frameshift vs. missense; data not shown) or CNV (amplification vs. deletion; data not shown). Of note, some discrepancies in mutation detection may have stemmed from the difference in specific genetic regions covered by each vendor due to the difference in target capture methods in hot spot amplicon designs. We could not assess this possibility because the region information was not available for some vendors offering hotspot panels.
There were differences in reporting when discrepancy in variant detection was reported (Table 3).
| Variant | Sample | Foundation One | Strand Advantage | GeneTrails | CANCP |
|---|---|---|---|---|---|
| APC p.Arg232X | 80 | y | y | NA | n |
| SMO p.Ala601Val | 240 | y | y | NA | n |
| TP53 p.Trp53X | 241 | y | y | y | n |
| TP53 p.Val122fsX26 | 84 | y | NA | y | n |
| AURKA amplification | 240 | n | y | NA | NA |
| EGFR amplification | 240 | n | y | n | NA |
| FGFR1 amplification | 358 | n | y | NA | NA |
| FGFR2 amplification | 358 | n | y | NA | NA |
| GNAS amplification | 26 | y | NA | n | NA |
| JAK2 amplification | 241 | n | y | NA | NA |
| KRAS amplification | 241 | n | y | n | NA |
| MET amplification | 202 | y (eq.) | y | n | NA |
| MET amplification | 358 | n | y | NA | NA |
| MYC amplification | 241 | n | y | NA | NA |
| NTRK1 amplification | 358 | n | y | NA | NA |
| SMO amplification | 358 | n | y | NA | NA |
| TP53 deletion | 80 | n | n | y | NA |
| TP53 deletion | 202 | n | n | y | NA |
| TP53 deletion | 289 | n | n | y | NA |
| TSC2 deletion | 289 | n | n | y | NA |
| BRCA2 deletion | 358 | n | y | NA | NA |
Table 3: Discrepantly detected variants.
StrandAdvantage didn’t test sample #26 and #84. GeneTrails didn’t test sample #358. CANCP didn’t perform CNV analyses. Other events marked as NA were not assessed because the specific target was not in the panel. This table includes variants reported as non-actionable or VUS.
These involved 4 mutations (APC p.Arg232X, SMO p.Ala601Val, TP53 p.Trp53X, and TP53 p.Val122fsX26). In all four cases, CANCP was the only vendor that didn’t report the mutations. For CNV, discrepancy involved amplification of 11 loci (AURKA, EGFR, FGFR1, FGFR2, GNAS, JAK2, KRAS, MET, MYC, NTRK1, and SMO) and deletion of 3 loci (TP53, BRCA2, and TSC2). GeneTrails was most often the only vendor for reporting deletions (four events), followed by StrandAdvantage (one event). StrandAdvantage most frequently was the sole vendor reporting amplifications (11 events), followed by Foundation one (three events). Interestingly, one amplification event was reported as “amplification” by StrandAdvantage, “equivocal amplification” by Foundation one, and not reported by GeneTrails. These findings strongly suggest that detection discrepancy depended on difference in definition of reportable CNV and analytical sensitivity setting, rather than technical error. An independent evaluation of discrepant variants on the studied samples was not performed by us.
Test-to-test comparison of actionable variant annotation and detection
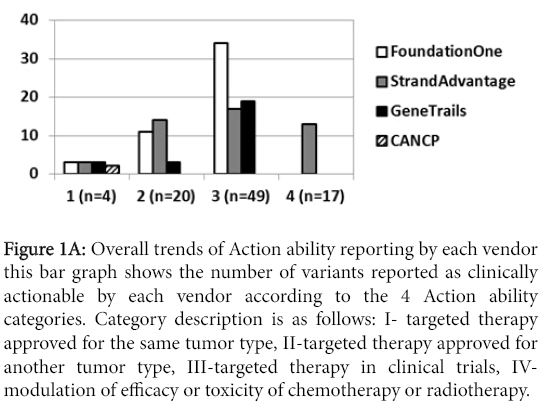
The number of clinically actionable variants reported greatly varied among the vendors. Figure 1A summarizes this number for each vendor.
Figure 1a: Overall trends of Action ability reporting by each vendor this bar graph shows the number of variants reported as clinically actionable by each vendor according to the 4 Action ability categories. Category description is as follows: I- targeted therapy approved for the same tumor type, II-targeted therapy approved for another tumor type, III-targeted therapy in clinical trials, IVmodulation of efficacy or toxicity of chemotherapy or radiotherapy.
However, simple comparison of reported variants may not be appropriate, since multiple factors may impact Action ability; a) error in detection or annotation, b) variant detection discrepancy due to analytical characteristics of assays, which were not independently validated in this study, c) different panel design and variant types assayed, and d) different clinical Action ability annotation. Furthermore, as was mentioned earlier, not all vendors could assess all samples due to sample exhaustion or incidents during shipment, which impacted numbers of the detected variants.
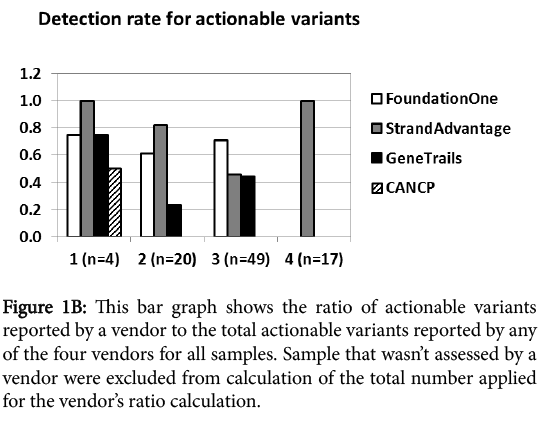
In order to eliminate the impact of samples that were not assessed by a specific vendor from this analysis, we calculated the Action ability detection rate of each vendor relative to total Action ability detected by all vendors in the samples that the vendor could report (Figure 1B).
Figure 1b: This bar graph shows the ratio of actionable variants reported by a vendor to the total actionable variants reported by any of the four vendors for all samples. Sample that wasn’t assessed by a vendor were excluded from calculation of the total number applied for the vendor’s ratio calculation.
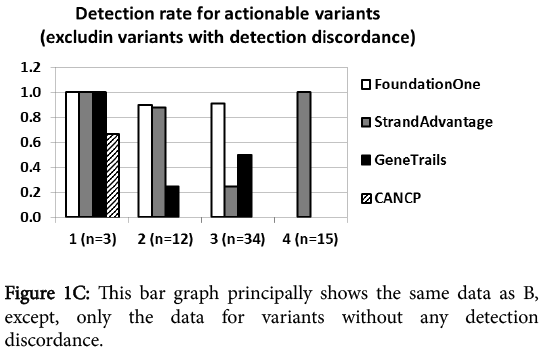
At first glance, category I Action ability (FDA-approved targeted therapy for the same tumor type) reporting was comparable for all vendors regardless of the panel size. CANCP reported actionable variants only in this category. Category II Action ability reporting (FDA-approved targeted therapy for other tumor types) was markedly higher for Foundation one and StrandAdvantage than GeneTrails. Foundation one had the highest report rate for category III Action ability (targeted therapy in clinical trial), followed by both StrandAdvantage and GeneTrails. Category IV Action ability (modulation of efficacy or toxicity of chemotherapy and radiotherapy) was reported only by StrandAdvantage. Figure 1C shows that this trend held true in the analysis excluding discrepantly detected variants (17 CNVs and 4 mutants described earlier), which should most directly reflect panel design and annotation/reporting policy difference.
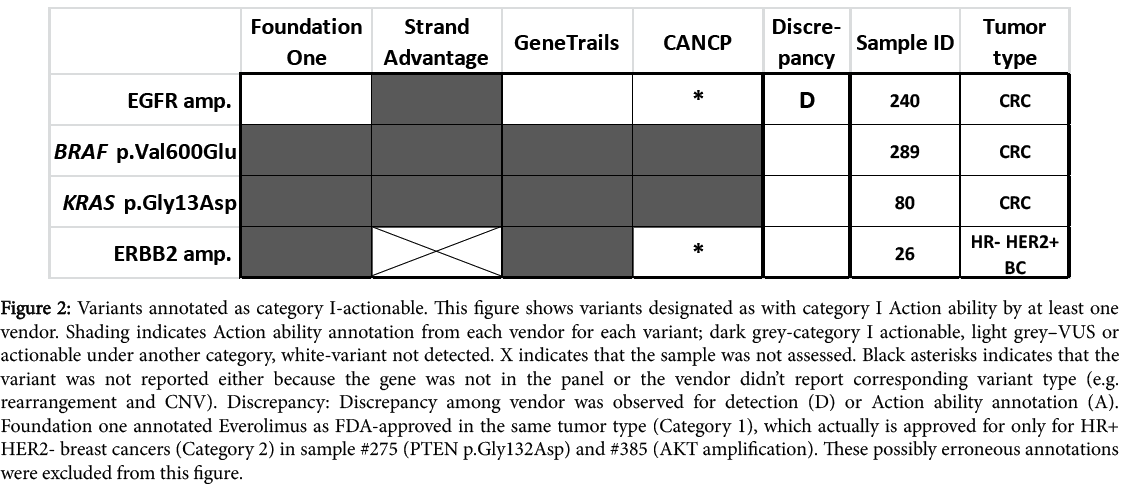
To better visualize differences in Action ability reporting between vendors and elucidate their potential causes, all clinically actionable variants reported in this study are listed according to the vendors and their status for each vendor (Figures 2-5 for Action ability categories IIV, respectively).
Figure 2: Variants annotated as category I-actionable. This figure shows variants designated as with category I Action ability by at least one vendor. Shading indicates Action ability annotation from each vendor for each variant; dark grey-category I actionable, light grey–VUS or actionable under another category, white-variant not detected. X indicates that the sample was not assessed. Black asterisks indicates that the variant was not reported either because the gene was not in the panel or the vendor didn’t report corresponding variant type (e.g. rearrangement and CNV). Discrepancy: Discrepancy among vendor was observed for detection (D) or Action ability annotation (A). Foundation one annotated Everolimus as FDA-approved in the same tumor type (Category 1), which actually is approved for only for HR+ HER2- breast cancers (Category 2) in sample #275 (PTEN p.Gly132Asp) and #385 (AKT amplification). These possibly erroneous annotations were excluded from this figure.
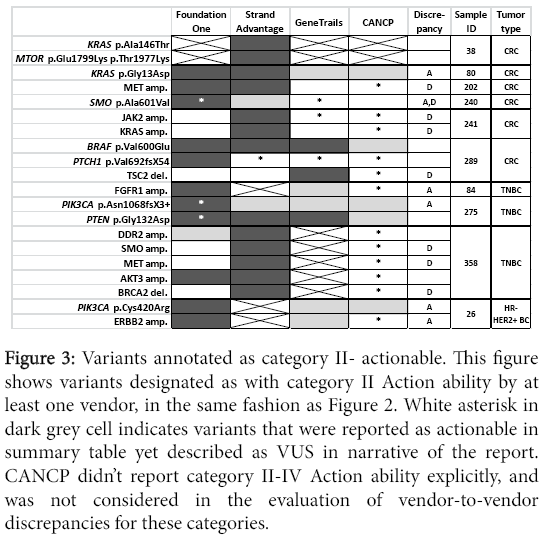
Figure 3: Variants annotated as category II- actionable. This figure shows variants designated as with category II Action ability by at least one vendor, in the same fashion as Figure 2. White asterisk in dark grey cell indicates variants that were reported as actionable in summary table yet described as VUS in narrative of the report. CANCP didn’t report category II-IV Action ability explicitly, and was not considered in the evaluation of vendor-to-vendor discrepancies for these categories.
As was noted earlier in Results, StrandAdvantage reports several amplifications which are not reported by other vendors, while GeneTrails reports a few deletions which are not reported by other vendors.
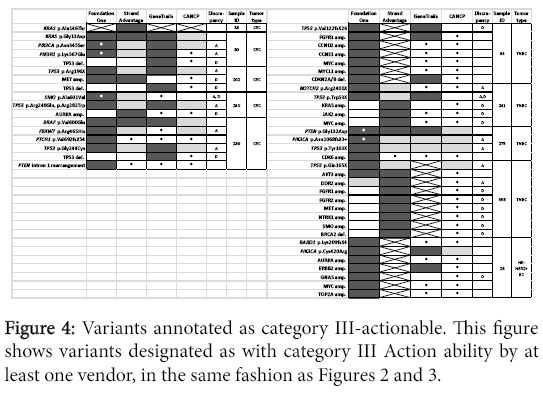
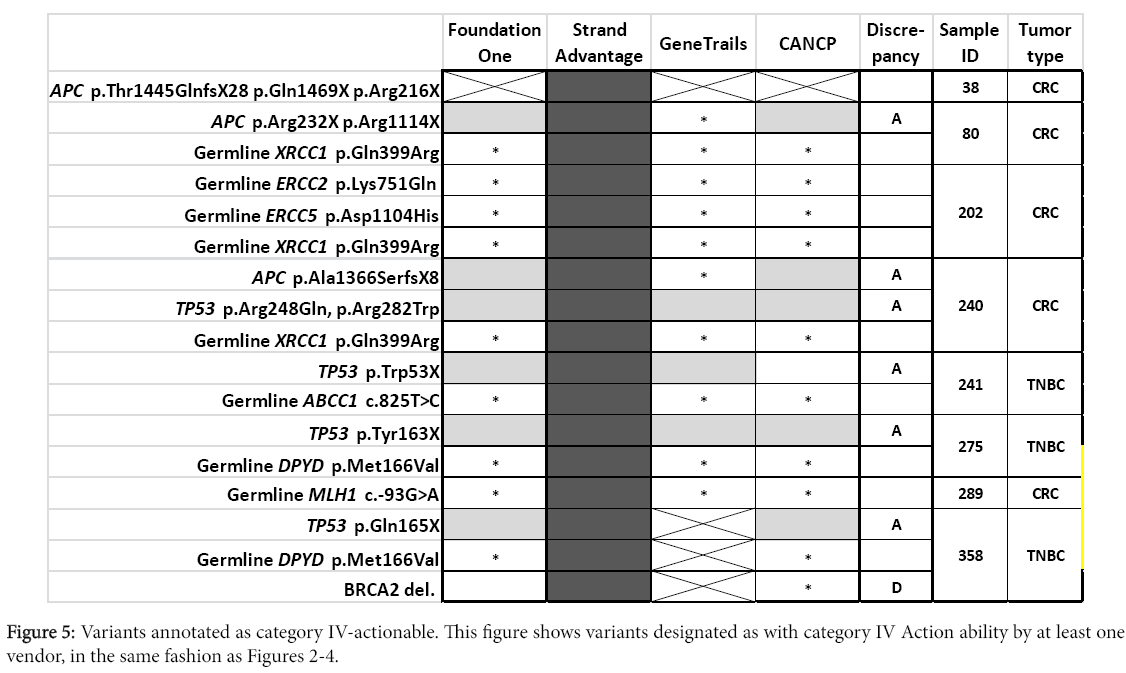
Larger panel design provided more actionable variants in category III (Foundation one and StrandAdvantage; (Figure 4) or category IV (StrandAdvantage; (Figure 5). Six variants were reported in targets assessed only by Foundation one (mutation of PTCH1 and BARD1, CNV of CDK6, MYCL1, and CCND2, as well as PTEN intron 1 rearrangement). The report also indicated possible targeted therapy clinical trials (except for PTCH1, where targeted therapy was approved for another tumor type). Six variants were reported for targets assessed only by StrandAdvantage (germline mutation of ERCC2. ERCC5, XRCC1, ABCC1, DPYD, and MLH1) which indicated either sensitivity or toxicity to chemotherapy.
In addition to the already described factors (i.e. difference in panel design and CNV detection sensitivity, or absence of category II-IV Action ability reporting by CANCP), discrepancies in Action ability reporting among vendors was likely dependent on differences in annotation. Major difference in annotation was observed for a total of 21 variants (including two occasions of double mutations of a gene in the same sample) corresponding to 12 genes (APC, DDR2, ERBB2, FBXW7, FGFR1, KRAS, NOTCH2, PIK3CA, PIK3R1, SMO, TP53). Distribution of different Action ability annotation was as follows: seven for category II (Figure 3), 13 for category III (Figure 4, including one occasion of double mutations in a single gene), and eight for category IV (Figure 5, including one occasion of double mutations in a single gene). No differences in action ability annotation were observed for category I.
Most differences in annotation appeared to depend on annotation focus and principles. This topic has been summarized in introduction and will be discussed further in Discussion. However, there were several occasions where either format of the report or an error seemed to contribute to reporting differences. GeneTrails’s free-text format reports didn’t always state specific drug name or FDA-approved tumor types, and often referred to off-label use of FDA-approved drugs under clinical trials. This reporting format may have attributed to the lack of category II annotation by GeneTrails (Figure 3) for KRAS p.Gly13Asp (sample #80), PIK3CA p.Cys420Arg (sample #26), ERBB2 amplification (sample #26), and FGFR1 amplification (sample #84). For variants SMO p.Ala601Val, PIK3CA p.Asn345Ser, and PIK3CA p.Asn1068fsX3+, Foundation one reported clinical trial availability in the summary, yet in the narrative stated that “mutation reported here has not been characterised.” This inconsistency within the report, possibly error, is likely the source of annotation discordance for these variants, since other vendors labelled these variants as VUS (Figure 4). Additionally, Foundation one annotated Everolimus as FDA-approved in the same tumor type (Category I) for two TNBC cases (#275, with PTEN p.Gly132Asp, and #385, with AKT amplification; Table 2), when it was approved only for HR+ HER2- breast cancers (Category II). This potential error was corrected prior to Action ability annotation comparison by us, as was stated under the Characteristics of reports, thus it is not reflected in Figures.
Discussion
The challenges in identifying actionable variants have been highlighted by various groups [14,15]. It is known that only certain ‘driver’ mutations are of consequence in tumorigenesis. Such driver mutations are usually shown to be functional in in vitro assays where oncogenic potential or activation of downstream pathways is observed. In the absence of this information, frequent occurrence in tumors may be taken as a surrogate. In the absence of either line of evidence, this inference may be speculative. Further, even if a variant is indeed confirmed to be oncogenic, its connection to therapy may be indirect and reliant on activation of a certain pathway. Even further, such pathway activation may be confounded by the presence of additional variants that modulate the pathway, which must be included in the assessment in vivo . Finally, reporting of Action ability requires considerations of practical matters, such as the handling of unapproved drugs and “off-label” use of approved drugs. Off-label use of drug is allowed when appropriate, but patient may encounter issues with reimbursement or access to pertinent clinical trials [16,17]. Similarly, recommendation of clinical trials where preliminary safety data have yet to be established (i.e. phase I) may require extra caution.
In the current study, there were multiple variants which are found by at least two vendors, but reported with differing clinical Action ability. As was mentioned in Results, CANCP did not explicitly report any Action ability other than FDA-approved targeted therapy in same tumor type. Some variants involved major discordance (i.e. presence versus absence of actionable variants in a category), and the others involved minor difference in recommendation within the category.
Major discordance
Major discordance involves Action ability annotation at category level. This type of discordance was observed for samples with variants of the TP53 and APC genes, NOTCH2 p.Arg2400X, FBXW7 p.Arg465His, PIK3CA p.Asn345Ser, PIK3R1 p.Lys567Glu, PTEN p.Gly132Asp, and DDR2 amplification. Our review revealed a few key contributing factors for this discordance: a) different functional significance annotation, due to different choices made at multiple decision points described above (in general, GeneTrails was the most aggressive, and CANCP was the most conservative), b) subtle difference in vendor’s field of focus (i.e. StrandAdvantage was almost the only vendor reporting Action ability on chemotherapy modulation), and c) handling of unapproved drugs and off-label use of approved drugs in the absence of clinical trials at phase II or later (Foundation one and GeneTrails made recommendation, while StrandAdvantage did not). Also, Foundation one noted impact of cofounding mutation/variant (e.g. HR status for Everolimus, oncogene mutation for FBX7 mutant targeting mTOR inhibitor use) in targeted therapy but did not mention it in the summary recommendation. Each of these variants is discussed in depth below with emphasis on differences in interpretation between vendors.
TP53 variants
TP53 somatic mutations were frequently detected by all vendors, but reported clinical Action ability markedly differed. Foundation one and GeneTrails referred to clinical trials, while StrandAdvantage only mentioned chemotherapy sensitivity/toxicity specific to the loss-offunction variant, p.Arg282Trp, with some reservation (sample #240, TNBC). All clinical trials reported by Foundation one and GeneTrails were phase I. StrandAdvantage cited indirect evidences for it’s claim: TP53 p.Arg282Trp mutation was associated with shorter overall survival in a mixed cancer patient population, as well as with possible resistance to cytochrome 450 metabolized drugs in cancer cell lines [18].
APC variants
APC somatic mutations were frequently detected by all vendors (Foundation one, StrandAdvantage, and CANCP) that tested APC in their panel [19]. However, StrandAdvantage is the only vendor that attributes these variants (truncation mutations in exon 14 or 16) to poor response to chemotherapy. Supportive evidence for this claim included a clinical study of CRC patients that suggested an association between nonsense or frameshift mutations in exon 16 of APC and poor response to 5-fluorouracil [20], with additional preclinical evidence to this effect.
Other variants with major discordance
The NOTCH2 truncation variant p.Arg2400X was detected in sample #241 by Foundation one and StrandAdvantage. It is a truncating mutation in a gene that behaves both as a tumor suppressor and as an oncogene [21], and has been reported as an indicator of potential response to Notch inhibitors (in clinical trial) by Foundation one and as non-actionable by StrandAdvantage. StrandAdvantage didn’t provide explanation for this designation, indicating that it annotated this variant as VUS. Foundation one cited a study of this variant showing its in vitro NOTCH2 activating affect as well as its cooccurrence with NOTCH2 copy number gain in lymphoma patients [22]. All clinical trials mentioned by Foundation one were at phase I, and no clinical study data are available at present for NOTCH2 inhibitor effectiveness in breast cancer patients.
The missense variant FBXW7 p.Arg465His in sample #289 was detected by Foundation one, Strandadvantage, and CANCP. It was deemed as actionable with clinical trials by Foundation one and nonactionable by StrandAdvantage. The referenced clinical trials were at phase I or II. Both vendors mentioned preclinical effectiveness of mTOR inhibitor therapy against FBXW7 variants [23]. Foundation one further supported this claim with a clinical case report [24], while StrandAdvantage dismissed the Action ability in this particular patient based on a clinical study showing that concomitant KRAS mutation mitigates the effectiveness of mTOR inhibitor therapy targeting FBXW7 variants [25,26]. CANCP didn’t report any clinical trials, but cited the same study and mentioned the negative interaction by coexisting mutations in oncogenes, such as KRAS.
The PIK3CA p.Asn345Ser and PIK3R1 p.Lys567Glu variants were detected in sample #80 by all vendors that assessed these genes (CANCP only analysed PIK3R1). However, they were listed as functional only by GeneTrails (Foundation one listed this PIK3CA variant as actionable in summary but as VUS in narrative-an error mentioned in Results). StrandAdvantage didn’t provide literature evidence for this annotation. CANCP stated the research on the PIK3CA variant was insufficient to ascertain its functionality. GeneTrails cited no reference for PIK3R1 variant, but stated that another mutant at the PIK3CA codon 345 demonstrated in vitro tumorigenicity [27]. This study was cited by Foundation one, too. Foundation one also stated that the detected PIK3CA and PIK3R1 missense mutations were located within domains of interaction between PIK3R1 and PIK3CA to negatively regulate the Akt pathway [28]. Foundation one also acknowledged that PIK3R1 p.Lys567Glu heterozygous cells was shown to be non-tumorigenic in vitro [29] and that direct evidence was missing for oncogenicity of PIK3CA p.Asn345Ser. The clinical trials recommended by GeneTrails were at phase I or II.
The PTEN p.Gly132Asp variant was detected in sample #275 by all vendors. Foundation one and CANCP regarded it non-functional (Foundation one report had summary-narrative inconsistency for this variant; while summary stated this variant to be actionable narrative reffered to it as functionally unclear). GeneTrails and StrandAdvantage designated this variant as actionable with approved drug for another tumor type in clinical trials. Foundation one, CANCP, and GeneTrails didn’t provide evidence for the functional significance of this missense variant. StrandAdvantage provided indirect support: in vitro data of PTEN functional loss caused by missense mutations at neighbouring codons (codons 121-131, and 134; [30,31]), and in-silico functional prediction results (Strand’s internal data). Referenced trials were phase I or II. StrandAdvantage cited clinical studies that showed mTOR inhibitor effectiveness in human cancers both dependent [32] and independent [33] of tumor PTEN-status, in addition to a few preclinical studies.
DDR2 amplification was detected in sample #358 by Foundation one and StrandAdvantage, but only StrandAdvantage reported it as clinically actionable. Foundation one designated this variant as VUS with no further description. StrandAdvantage stated that, while direct evidence for DDR2 amplification was lacking, DDR2 activation was shown to be oncogenic in a few preclinical studies of DDR2 activating mutation [34-36]. Similarly, the response to tyrosine kinase inhibitor therapy in patients with DDR2 activating mutations was referenced as a supportive evidence [34,37,38]. The recommended clinical trial was at phase II.
Minor discordance (intra-category discrepancy)
An example of minor discrepancy in recommendation between vendors was seen for KRAS p.Gly13Asp mutation in a CRC sample #80. All vendors report resistance to cetuximab and panitumumab (both category I) for this sample, but only StrandAdvantage noted additional detail that this variant may be more responsive to these drugs than other KRAS codon 12/13 mutants, based both clinical and preclinical studies [39-41]. Also, only StrandAdvantage reported resistance to VEGF inhibitor Bevacizumab (category I) for this variant based on meta-analysis of 12 clinical trial data [42]. Further, a category II recommendation for this variant differed between Foundation one (Trametinib) versus StrandAdvantage (Sorafenib). Sarafenib was reported by StrandAdvantage to have clinical evidence specifically in CRC as a second or later-line treatment [43], while clinical effectiveness of Trametinib was reported in melanoma [44] and other solid tumors [45,46], but not yet specifically in CRC.
Conclusions
Our comparison between the 4 vendors shows substantial commonality as well as differences of commercial NGS-based gene panel analysis of solid tumor in panel design, reporting policies, and clinically actionable information provided. While the study was too small to derive statistically significant inferences, some qualitative trends were observed. All enrolled tests, regardless of the panel design, were capable of reporting well-established actionable variants (i.e. those with FDA-approved target therapy for the specific tumor type) with consistent clinical interpretations. In contrast, reporting of other Action ability was heavily impacted by panel design, reporting policy, and annotation policy.
In general, the larger panels (Foundation one and StrandAdvantage) detected and reported more actionable variants besides targeted therapies approved for the specific tumor type than the remaining smaller panels. Of the latter, GeneTrails covered clinical trial Action ability well, while CANCP massively limited its actionable variant detection capability by not reporting CNVs and not referring to specific clinical trials. Foundation one reported more genes and rearrangements with targeted therapy in trials, particularly in early phase trials with novel investigational drugs, compared the other panels. StrandAdvantage provided information on variant modulation of chemotherapy efficacy and toxicity by germline and somatic variants, which the other vendors didn’t provide. StrandAdvantage
reported relatively less clinical trial Action ability for its expansive panel size, in part due to its annotation policy that is further described in the next paragraph. Of note, the assessed tumor types were not those in which gene rearrangements have large impact on Action ability (e.g. non-small cell lung cancer). Therefore, the current study is unsuited for full evaluation of the utility of the Foundation one and StrandAdvantage panels’ rearrangement targets.
Based on the examination of variants with different interpretations several important distinctions were noted in policies for clinical trials reporting: decision to report (Foundation one and GeneTrail) or not (StrandAdvantage) variants when only phase I trials are available; decision to report off-label recommendations in the absence of a trial relevant to the tumor type (Foundation one) or not (StrandAdvantage); decision to report Action ability supported only by preclinical data (Foundation one) or not (StrandAdvantage and GeneTrails). CANCP didn’t report availability of specific clinical trial at all. Accordingly, StrandAdvantage was generally more conservative than other vendors in reporting available clinical trials, thus their coverage of clinical trials was relatively small for its panel size.
Analytical sensitivity/specificity of mutation detection appeared comparable among vendors, while CNV reporting was highly variable. The difference in reporting accounted for significant differences in provided clinically actionable information. StrandAdvantage detected more clinically actionable amplifications, while GeneTrails detected more clinically actionable deletions than the other vendors. It should be noted that the veracity of CNV calls was not independently verified in our study. Vendors are encouraged to perform this validation on this own. It may also be desirable to develop a consensus in the definition of clinically impactful CNV.
Report formats of Foundation one and StrandAdvantage were the most comprehensive with statement of drug FDA-approval status, clinical trial description, and references to supporting literature. Both vendors provided concise summary for convenience, but caution should nonetheless be exercised for occasional discrepancy between summary and narrative (observed in Foundation one). GeneTrails and CANCP had shorter report formats. GeneTrails didn’t provide literature information, thus verification of the recommendation required independent literature search. CANCP mentioned ongoing research with literature but without information on availability of a specific drug or a clinical trial, requiring active literature/database search to take full advantage of the variant data.
All the vendors continue to improve their reports and gene panel contents, so the trends reported in our study, may change. It is clear though that sequencing methods, choice of genes, bioinformatics methods, and variant curation and interpretation policies will have a major bearing on the accuracy and nature of reporting. Larger studies will be needed to quantify the qualitative differences outlined above and evaluate the benefit to the patient. Such rigorous analysis will no doubt improve the practice of precision medicine in the future.
References
- Lièvre A, Bachet JB, Boige V, Cayre A, Le Corre D, et al. (2008) KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol 26: 374-379.
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, et al. (2008) Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 26: 1626-1634.
- Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, et al. (2013) Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 369: 1023-1034.
- Chen HJ, Mok TS, Chen ZH, Guo AL, Zhang XC, et al. (2009) Clinicopathologic and molecular features of epidermal growth factor receptor T790M mutation and c-MET amplification in tyrosine kinase inhibitor-resistant Chinese non-small cell lung cancer. Pathol Oncol Res 15: 651-658.
- Arcila ME, Oxnard GR, Nafa K, Riely GJ, Solomon SB, et al. (2011) Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res 17: 1169-1180.
- Kishiki T, Ohnishi H, Masaki T, Ohtsuka K, Ohkura Y, et al. (2014) Overexpression of MET is a new predictive marker for anti-EGFR therapy in metastatic colorectal cancer with wild-type KRAS. Cancer Chemother Pharmacol 73: 749-757.
- Bean J, Brennan C, Shih JY, Riely G, Viale A, et al. (2007) MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. ProcNatlAcad Sci USA 104: 20932-20937.
- Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, et al. (2010) Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 17: 77-88.
- Song N, Liu S, Zhang J, Liu J, Xu L, et al. (2014) Cetuximab-induced MET activation acts as a novel resistance mechanism in colon cancer cells. Int J Mol Sci15: 5838-5851.
- Mohan S, Heitzer E, Ulz P, Lafer I, Lax S, et al. (2014) Changes in colorectal carcinoma genomes under anti-EGFR therapy identified by whole-genome plasma DNA sequencing. PLoS Genet 10: e1004271.
- Bokemeyer C, Van Cutsem E, Rougier P, Ciardiello F, Heeger S, et al. (2012) Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur J Cancer 48: 1466-1475.
- Wang Q, Hu WG, Song QB, Wei J (2014) BRAF V600E mutation as a predictive factor of anti-EGFR monoclonal antibodies therapeutic effects in metastatic colorectal cancer: a meta-analysis. Chin Med Sci J 29: 197-203.
- Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, et al. (2013) Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31: 1023-1031.
- Good BM, Ainscough BJ, McMichael JF, Su AI, Griffith OL (2014) Organizing knowledgeto enable personalization of medicine in cancer. Genome Biol 15: 438.
- Dienstmann R, Dong F, Borger D, Dias-Santagata D, Ellisen LW, et al. (2014) Standardized decision support in next generation sequencing reports of somatic cancer variants. MolOncol 8: 859-873.
- Beck JM, Azari ED (1998) FDA, off-label use, and informed consent: debunking myths and misconceptions. Food Drug L J 53: 71-104
- Laetz T, Silberman G (1991) Reimbursement policies constrain the practice of oncology. JAMA266: 2996-2999.
- Xu J, Wang J, Hu Y, Qian J, Xu B, et al. (2014) Unequal prognostic potentials of p53 gain-of-function mutations in human cancers associate with drug-metabolizing activity. Cell Death Dis 5: e1108.
- Lehmann-Che J, André F, Desmedt C, Mazouni C, Giacchetti S, et al. (2010) Cyclophosphamide dose intensification may circumvent anthracycline resistance of p53 mutant breast cancers. Oncologist 15: 246-252.
- Chen SP, Wu CC, Lin SZ, Kang JC, Su CC, et al. (2009) Prognostic significance of interaction between somatic APC mutations and 5-fluorouracil adjuvant chemotherapy in Taiwanese colorectal cancer subjects. Am J Clin Oncol 32: 122-126.
- Lobry C, Oh P, Aifantis I (2011) Oncogenic and tumor suppressor functions of Notch in cancer: it's NOTCH what you think. J Exp Med208: 1931-1935
- Lee SY, Kumano K, Nakazaki K, Sanada M, Matsumoto A, et al. (2009) Gain-of-function mutations and copy number increases of Notch2 in diffuse large B-cell lymphoma. Cancer Sci 100: 920-926.
- Mao JH, Kim IJ, Wu D, Climent J, Kang HC, et al. (2008) FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science 321: 1499-1502.
- Villaruz LC, Socinski MA (2014) Temsirolimus therapy in a patient with lung adenocarcinoma harboring an FBXW7 mutation. Lung Cancer 83: 300-301
- Jardim DL, Wheler JJ, Hess K, Tsimberidou AM, Zinner R, et al. (2014) FBXW7 mutations in patients with advanced cancers: clinical and molecular characteristics and outcomes with mTOR inhibitors. PLoS One 9: e89388.
- Di Nicolantonio F, Arena S, Tabernero J, Grosso S, Molinari F, et al. (2010) Deregulation of thePI3K and KRAS signaling pathways in human cancer cells determines their responseto everolimus. J Clin Invest 120: 2858-2866.
- Gymnopoulos M, Elsliger MA, Vogt PK (2007) Rare cancer-specific mutations in PIK3CAshow gain of function. ProcNatlAcad Sci USA 104: 5569-5574.
- Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, et al. (2007) The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science 318: 1744-1748.
- Cheung LW, Hennessy BT, Li J, Yu S, Myers AP, et al. (2011) High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov 1: 170-185.
- Han SY, Kato H, Kato S, Suzuki T, Shibata H, et al. (2000) Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer Res 60: 3147-3151.
- Rodríguez-Escudero I, Oliver MD, Andrés-Pons A, Molina M, Cid VJ, et al. (2011) A comprehensive functional analysis of PTEN mutations: implications in tumor- and autism-related syndromes. Hum Mol Genet 20: 4132-4142.
- Gonzalez-Angulo AM, Blumenschein GR Jr (2013) Defining biomarkers to predict sensitivity to PI3K/Akt/mTOR pathway inhibitors in breast cancer. Cancer Treat Rev 39: 313-320
- Oza AM, Elit L, Tsao MS, Kamel-Reid S, Biagi J, et al. (2011) Phase II study of temsirolimus in women with recurrent or metastatic endometrial cancer: a trial of the NCIC Clinical Trials Group. J Clin Oncol 29: 3278-3285.
- Hammerman PS, Sos ML, Ramos AH, Xu C, Dutt A, et al. (2011) Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov 1: 78-89.
- Olaso E, Labrador JP, Wang L, Ikeda K, Eng FJ, et al. (2002) Discoidin domain receptor 2 regulates fibroblast proliferation and migration through the extracellular matrix in association with transcriptional activation of matrix metalloproteinase-2. J BiolChem 277: 3606-3613.
- Zhang K, Corsa CA, Ponik SM, Prior JL, Piwnica-Worms D, et al. (2013) The collagen receptor discoidin domain receptor 2 stabilizes SNAIL1 to facilitate breast cancer metastasis. Nat Cell Biol 15: 677-687.
- Day E, Waters B, Spiegel K, Alnadaf T, Manley PW, et al. (2008) Inhibition of collagen-induced discoidin domain receptor 1 and 2 activation by imatinib, nilotinib and dasatinib. Eur J Pharmacol 599: 44-53.
- Pitini V, Arrigo C, Di Mirto C, Mondello P, Altavilla G (2013) Response to dasatinib in a patient with SQCC of the lung harboring a discoid-receptor-2 and synchronous chronic myelogenous leukemia. Lung Cancer 82: 171-172.
- De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, et al. (2010) Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 304:1812-1820.
- Tejpar S, Celik I, Schlichting M, Sartorius U, Bokemeyer C, et al. (2012) Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J Clin Oncol 30: 3570-3577.
- Kumar SS, Price TJ, Mohyieldin O, Borg M, Townsend A, et al. (2014) KRAS G13D Mutation and Sensitivity to Cetuximab or Panitumumab in a Colorectal Cancer Cell Line Model. Gastrointest Cancer Res 7: 23-26.
- Petrelli F, Coinu A, Cabiddu M, Ghilardi M, Barni S (2013) KRAS as prognostic biomarker in metastatic colorectal cancer patients treated with bevacizumab: a pooled analysis of 12 published trials. Med Oncol30: 650
- Samalin E, Bouché O, Thézenas S, Francois E, Adenis A, et al. (2014) Sorafenib and irinotecan (NEXIRI) as second- or later-line treatment for patients with metastatic colorectal cancer and KRAS-mutated tumours: a multicentre Phase I/II trial. Br J Cancer 110: 1148-1154.
- Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, et al. (2012) Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 367: 107-114.
- Infante JR, Fecher LA, Falchook GS, Nallapareddy S, Gordon MS, et al. (2012) Safety, pharmacokinetic, pharmacodynamic, and efficacy data for the oral MEK inhibitor trametinib: a phase 1 dose-escalation trial. Lancet Oncol 13: 773-781.
- Infante JR, Papadopoulos KP, Bendell JC, Patnaik A, Burris HA 3rd, et al. (2013) A phase 1b study of trametinib, an oral Mitogen-activated protein kinase kinase (MEK) inhibitor, in combination with gemcitabine in advanced solid tumours. Eur J Cancer 49: 2077-2085.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 12867
- [From(publication date):
August-2016 - Nov 21, 2024] - Breakdown by view type
- HTML page views : 11992
- PDF downloads : 875