Research Article Open Access
miR-143 is Associated with ER-�?±36 Expression Gastric Carcinoma of Xenografted Model
Zou Feng1, Zhang Shangkun1,2, Li Yan1, Chen Ying1 and Liu Lijiang1*1 Department of Pathology and Pathophysiology, School of Medicine, Jianghan University, Wuhan, China
2JiangDa Pathology Institution, Wuhan 430056, Hubei Province, China
- Corresponding Author:
- Liu Lijiang
Department of Pathology and Pathophysiology
School of Medicine, Jianghan University, Wuhan, China
E-mail: liulijiang@163.com
Received date September 28, 2013; Accepted date November 11, 2013; Published date November 18, 2013
Citation: Feng Z, Shangkun Z, Yan L, Ying C, Lijiang L (2013) miR-143 is Associated with ER-a36 Expression Gastric Carcinoma of Xenografted Model. J Biomim Biomater Tissue Eng 18:117. doi:10.4172/1662-100X.1000117
Copyright: © 2013 Feng Z, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biomimetics Biomaterials and Tissue Engineering
Abstract
We investigated the expression profile of microRNA (miRNA) in ER-α36-mediated transplantable tumours. SGC7901 cell lines of higher or lower ER-α36 expression were constructed with a lentivirus vector. Five million SGC7901 cells were injected into nude mice to form a gastric xenografted carcinoma in the subcutaneous tissue of nude mice. Observation was performed for 30 days. The weight and propagation of the tumours were determined. miRNA expression profiles of the three SGC7901 cell lines were analysed by miRNA microarray. miR-143 expression in transplantable tumours was determined by real-time PCR using TaqMan assay. The expression level of ER-α36 was proportional to the size and propagation of gastric xenografted carcinomas in nude mice. The expression level of miR-143 significantly decreased in highly expressing ER-α36 SGC7901 cells and significantly increased in lowly expressing ER-α36 SGC7901 cells. miR-143 expression results in transplantable tumours amongst nude mice were consistent with the miRNA microarray findings. The expression of miR-143 was suppressed by high ER-α36 expression. These findings suggest that deregulation of miR-143 plays an important role in the progression of ER-α36-mediated gastric xenografted carcinoma.
Keywords
Gastric cancer; ER-α36; miR-143; SGC7901; Gastric xenografted carcinoma
Introduction
The diverse activities of oestrogens and anti-oestrogens mediated by specific receptors designated as oestrogen receptors (ERs) are well documented. Two ERs, ER-α and ER-β, which are ligand-activated transcription factors that stimulate target gene transcription, have been identified. Two forms of ER-α, a full-length form (66 kDa) and a short-length one (46 kDa), have been found on or near the plasma membrane and mediate membrane-initiated oestrogen signalling [1,2]. In 2005, a novel variant of ER-α with a molecular weight of 36 kDa was identified and cloned; this variant was called ER-α36. ER-α36 differs from the original 66 kDa ER-α (ER-α66) in that it lacks two transcriptional activation domains (AF-1 and AF-2) but retains the DNA-binding and partial dimerisation and ligand-binding domains. ER-α36 is predominantly expressed in the plasma membrane of breast cancer cells (ER-positive and -negative), mediates membrane-initiated oestrogen and anti-oestrogen signalling, such as activation of the MAPK/ERK and PI3K/AKT signalling pathways, and stimulates cell proliferation [3-5].
In our previous studies, we examined the expression and localisation of ER-α36 in established gastric cancer cell lines of SGC7901, BGC823, AGS, and MKN45. Our results show that ER-α36 is predominantly expressed in the plasma membrane of gastric cancer cell lines. We report the expression of ER-α36 mRNA in human gastric adenocarcinoma tissues in comparison with non-tumour mucosa. Our data suggest that upregulation of ER-α36 mRNA is significantly correlated with lymph node metastasis and infiltration of gastric carcinoma. The mechanisms of metastasis and infiltration of gastric carcinoma may be correlated with ER-α36 protein expression [6-9].
MicroRNAs (miRNAs) are small, endogenously conserved, noncoding RNAs ranging from 20 nt to 22 nt in length. miRNAs can induce mRNA translation inhibition or degradation through regulation of gene expression at the post-transcriptional level by binding to the 3'-UTR of target mRNAs [10,11]. This class of RNAs is believed to regulate about 30% of all human genes. Alterations in the expression of miRNAs are considered a common characteristic of all human tumours. MicroRNAs have been shown to be involved in a variety of tumour cell biological processes, including cell differentiation, proliferation, apoptosis, metastasis, and invasion [12].
MicroRNAs are frequently located near cancer-associated genomic regions or fragile sites and play a vital role as a novel class of oncogenes or tumour-suppressor genes [13]. Alterations in miRNA expression levels have been demonstrated in several gastrointestinal tumours, and miRNAs have been found to influence the prognostic significance of the tumours. Several miRNAs, such as miR-1, miR-20a, miR-27a, miR- 34, and miR-423-5p, can also regulate gene expression in gastric cancer [14,15]. Increasing miRNA expression levels have been associated with gastric cancer.
To the best of our knowledge, the correlation between miRNA expression and ER-α36 in gastric cancer has yet to be reported. In this study, we detected the miRNA expression of gastric xenografted carcinoma in nude mice and observed the miRNA expression of stable SGC7901 cell lines with lower or higher ER-α36 expression. The study aimed to investigate the relationship between the expressions of ER-α36 and miRNA in gastric cancer.
Materials and Methods
Cell culture
The human gastric adenocarcinoma cell line SGC7901 was obtained from the Chinese Academy of Medical Sciences Cell Centre of Basic Medicine (Beijing, China). Cell lines were cultured in RPMI 1640 containing 10% FCS at 37°C in a 5% CO2 atmosphere.
Construction of stable SGC7901 cell lines with lower or higher ER-α36 expression
ER-α36-silenced plasmids (PLKO.1-PURO-SP6-ER-α36-L) and ER-α36-overexpressing plasmids (PLJM1- ER-α36-H) were confirmed based on the effective sequences of siRNA targeting of the ER-α36 gene obtained from Wang Zhaoyi’s laboratory. The complementary DNA containing both sense and antisense oligo DNAs of the targeted sequence was designed using the following sequences: ER-α36F: CCGGTTGATGCCAATAGGTACTGAACTCGAGTTCAGTACC TATTGGCATCAATTTTTTG;ER-α36R:AATTCAAAAAA TTGATGCCAATAGGTACTGAACTCGAGTTCAGTACCTATTGG CATCAA. Stable SGC7901 cell lines with lower or higher ER-α36 expression were constructed by infection of PLKO.1-PURO-SP6-ER- α36-L and PLJM1- ER-α36-H after PCR confirmation and sequencing. ER-α36 low SGC7901 cells stably expressing PLKO.1-PURO-SP6- ER-α36-L (low ER-α36 group) and ER-α36 high SGC7901 cells stably expressing PLJM1-ER-α36-H (high ER-α36 group) were established using puromycin for isolation. The ER-α36 control SGC7901 group was constructed by transferring empty vector (PLKO.1-PURO-scremble shRNA) at the same time.
Western blot analysis
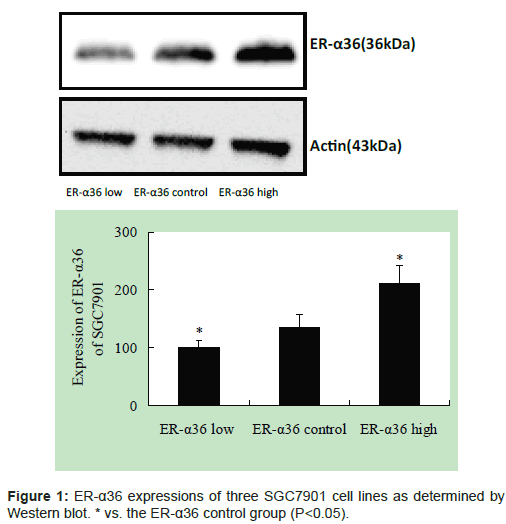
Cell lysates from SGC7901 cells were prepared in 1 mL lysis buffer (Sigma, USA) on ice in 1.5 mL microtubes. The lysates were solubilised by continuous stirring for 1 h at 4°C and then centrifuged for 10 min at 14000 × g. The supernatants were collected, and protein concentrations were measured using BCA protein assay reagent. Cell lysates were subjected to SDS-PAGE. After blotting, membranes were blocked with 5% skim milk in Tris-buffered saline containing 0.1% Tween 20 for 1 h and then incubated with primary antibodies for 16 h at 4°C. Primary antibodies included those against ER-α36 (Zhaoyi Wang’s laboratory, USA) and actin (Santa Cruz, USA). The membranes were incubated with anti-mouse IgG antibody for 1 h after three washes in Tris-buffered saline containing 0.1% Tween 20. The proteins were detected with an ECL system (KPL Amersham).
Nude mice experiments
Nine male SPF BALB/c-nu mice (20-25 g, 6 weeks old) were supplied by the Experimental Animal Centre of Hubei Province. The experiment was divided into three groups: the control SGC7901 group (ER-α36 control group, n=3), the SGC7901 with high ER- α36 expression group (high ER-α36 group, n=3), and the SGC7901 with low ER-α36 expression group (low ER-α36 group, n=3). Five million SGC7901 cells were injected into the nude mice to form a SGC7901 gastric xenografted carcinoma in the subcutaneous tissue, and observation was performed for 30 d. The sizes and weights of the tumours that developed were determined.
HE and immunohistochemical staining of gastric xenografted carcinoma
HE staining was performed according to the conventional method. Immunohistochemical staining was performed on 4 μm thick tumour sections via a two-step assay. Tissue slides were incubated overnight with the Ki-67 antibody diluted to 1:50 at 4°C in a humidified chamber. Ki-67 staining was scored as described by Couzin: positive were the Ki- 67 cell number when counted 100 cell in high power field (×400) [15].
MiRNA microarray of the three SGC7901 cell lines
Total RNA from the three SGC7901 cell lines was isolated using Trizol reagent (Invitrogen, USA). The concentration and quality of RNA were measured by a NanoDrop ND-1000 system at 260 and 280 nm (A260/280). RNA labelling and hybridisation were completed by BGI (Shenzhen, China) according to the manufacturer’s instructions. Only those with increases or decreases in expression greater than 2-fold relative to the control were considered significantly changed. Samples were clustered according to their miRNA profile using Cluster 3.0.
TaqMan miRNA real-time RT-PCR quantification of gastric xenografted carcinoma
Real-time RT-PCR analysis of miRNA expression was carried out using TaqMan microRNA assay kits according to the manufacturer’s protocol (Applied Biosystems, USA). Briefly, total RNA was extracted using an isolation additive buffer. Reverse transcriptase was synthesised from the total RNA (10 ng) using gene-specific primers. PCR reactions were run on a Step One Plus Real-Time PCR machine (Applied Biosystems) and analysed using Step One Plus System SDS software.
U6 small nuclear RNA (Applied Biosystems) was used as an internal control to normalise RNA input. The Ct value was defined as the fractional cycle number at which fluorescence exceeds the fixed threshold. Fold-changes in tumour expression relative to corresponding normal tissues were calculated using the 2-ΔΔCt method after normalisation to the endogenous control. All experiments were carried out in triplicate.
Statistical analysis
All values are expressed as mean ± SEM. Statistical significance was determined using ANOVA followed by t-tests. P<0.05 was considered statistically significant.
Results
Establishment of stable SGC7901 cell lines with lower or higher ER-α36 expression
The ER-α36 expression of three SGC7901 cell lines was determined by Western blot. This result confirms that construction of stable SGC7901 cell lines with lower and higher ER-α36 expression was successful (Figure 1).
Formation of distinct gastric xenografted carcinoma in nude mice by injection of the three SGC7901 cell lines
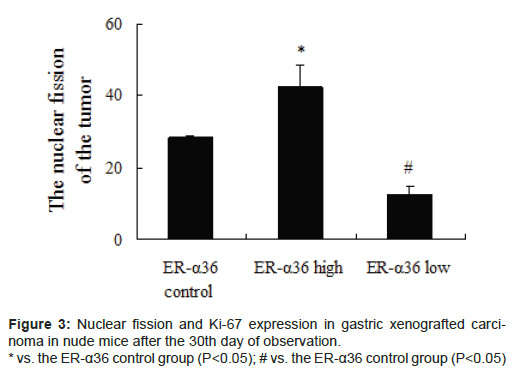
Gastric xenografted carcinoma was observed in all nude mice. After the 30th day, the tumour weight was highest in the high ER-α36 group, moderate in the control ER-α36 group, and lowest in the low ER-α36 group. Significant differences in tumour size were observed amongst groups (Figure 2, P<0.05). Nuclear fission and Ki-67 expression showed a similar trend amongst groups (Figure 3, P<0.05). ER-α36 expression was proportional to the size and propagation of gastric xenografted carcinoma.
Differential expression of miRNAs amongst the three SGC7901 cell lines
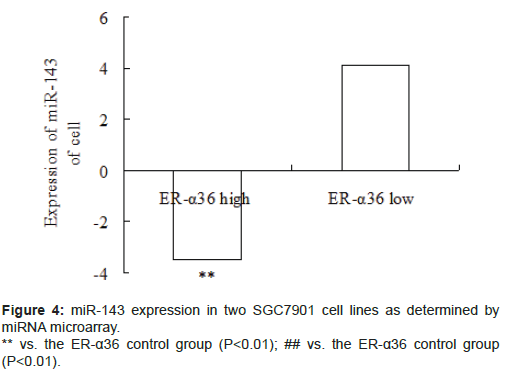
Several miRNAs were deregulated in ER-α36-mediated gastric cancer. The expression level of miR-143 significantly decreased in highly expressing ER-α36 SGC7901 cells and significantly increased in lowly expressing ER-α36 SGC7901 cells (Figure 4, P<0.01).
Expression of miR-143 in gastric xenografted carcinoma in nude mice
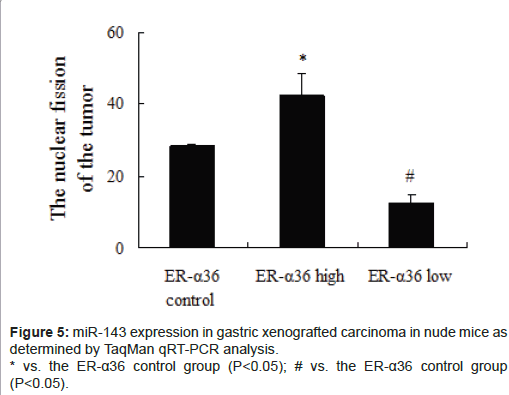
miR-143 expression levels in transplantable tumours in nude mice were significantly downregulated in the high ER-α36 group (Figure 5, P<0.05) and upregulated in the low ER-α36 group (Figure 5, P<0.05) compared with the control ER-α36 group. These results are consistent with those obtained from our miRNA microarray studies.
Discussion
ER-α36, a novel variant of ER-α, is identical in most aspects to the ER-α66 protein encoded by exons 2–6 of the ER-α66 gene. However, ER-α36 lacks intrinsic transcriptional activity but can suppress transactivation activities mediated by the AF-1 and AF-2 domains of liganded and unliganded ER-α66 and ER-α. These properties indicate that ER-α36 is a potent inhibitor of genomic oestrogen signalling. ER- α36 was first discovered by Wang in breast cancer and later reported in other cancers. The role of ER-α36 in gastric cancer remains unclear. In our previous study, we found that gastric cancer cells treated with 17β-oestradiol respond by the effects of ER-α36. Tamoxifen could inhibit the growth of BGC823 gastric cancer cells, and ER-α36 may transmit tamoxifen growth-regulated signals in gastric cancer via a membrane-based pathway. These results show that ER-α36 expression is associated with tumourigenesis.
Deregulation of miRNA expression in cancer has been widely reported. However, previous studies focused only on miRNAs expressed in tumour tissues and cells, and the specificity of biomarkers based on a single tumour-specific miRNA was generally poor. Not much is known about the involvement of miR-143 in cancer [16]. The tumour-suppressive effects of miR-143 in squamous cell carcinoma cell lines and in colorectal cancer have been previously reported [17,18]. In this study, our miRNA microarray results indicated that miR-143 is a potentially useful biomarker in gastric cancer, especially in ER-α36- mediated gastric cancer. In the next phase of our study, we will focus on the regulation of miR-143 target genes in gastric cancer.
In summary, this study determined that miRNA deregulation is associated with ER-α36-mediated gastric cancer. miR-143 expression is suppressed by high levels of ER-α36 in gastric cancer cells and gastric xenografted carcinoma in nude mice. Determination of whether or not miR-143 is regulated by oestrogen receptors will be an interesting subject for future research. We believe that miR-143 is involved in oestrogen receptor signal transduction. Our findings provide important insights into the progression of ER-α36-mediated gastric cancer.
Acknowledgement
We thank Prof. Zhaoyi Wang for providing the ER-α36 antibody used in this work. This work was supported grants awarded to National Natural Science Foundation of China (Grant No.81272754) and Wuhan Young Chenguang Foundation (Grant No.2013071004010475).
References
- Pedram A, Razandi M, Levin ER (2006) Nature of functional estrogen receptors at the plasma membrane. Mol Endocrinol 20: 1996-2009.
- Tokunaga A, Nishi K, Matsukura N, Tanaka N, Onda M, et al. (1986) Estrogen and progesterone receptors in gastric cancer. Cancer 57: 1376-1379.
- Wang Z, Zhang X, Shen P, Loggie BW, Chang Y, et al. (2006) A variant of estrogen receptor-{alpha}, hER-{alpha}36: transduction of estrogen- and antiestrogen-dependent membrane-initiated mitogenic signaling. Proc Natl Acad Sci U S A 103: 9063-9068.
- Zou Y, Ding L, Coleman M, Wang Z (2009) Estrogen receptor-alpha (ER-alpha) suppresses expression of its variant ER-alpha 36. FEBS Lett 583: 1368-1374.
- Wang Z, Zhang X, Shen P, Loggie BW, Chang Y, et al. (2005) Identification, cloning, and expression of human estrogen receptor-alpha36, a novel variant of human estrogen receptor-alpha66. Biochem Biophys Res Commun 336: 1023-1027.
- Hao D, Xuming W, Huang X, Zhen H, Chen Y, et al. (2011) ER-a36 participate growth inhibiting which induced by Tamoxifen in gastric cancer cell line BGC823. 2011 International Conference on Human Health and Biomedical Engineering, 1062-1065.
- Deng H, Zhen H, Fu Z, Huang X, Zhou H, et al. (2012) The antagonistic effect between STAT1 and Survivin and its clinical significance in gastric cancer. Oncol Lett 3: 193-199.
- Deng H, Huang X, Fan J, Wang L, Xia Q, et al. (2010) A variant of estrogen receptor-alpha, ER-alpha36 is expressed in human gastric cancer and is highly correlated with lymph node metastasis. Oncol Rep 24: 171-176.
- Wang XM, Liu JJ, Deng H, Chen Y, Liu LJ (2011) Effect of ER-a36 on the growth of the SGC-7901 in nude mice. Shijie Huaren Xiaohua Zazhi 19: 2919-2924.
- Lee Y, Kim M, Han J, Yeom KH, Lee S, et al. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051-4060.
- Wu L, Fan J, Belasco JG (2006) MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A 103: 4034-4039.
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, et al. (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 103: 2257-2261.
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, et al. (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 101: 2999-3004.
- Liu R, Zhang C, Hu Z, Li G, Wang C, et al. (2011) A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer 47: 784-791.
- Fasanella S, Leonardi E, Cantaloni C, Eccher C, Bazzanella I, et al. (2011) Proliferative activity in human breast cancer: Ki-67 automated evaluation and the influence of different Ki-67 equivalent antibodies. Diagn Pathol 6 Suppl 1: S7.
- Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, et al. (2009) miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 460: 705-710.
- Wu BL, Xu LY, Du ZP, Liao LD, Zhang HF, et al. (2011) MiRNA profile in esophageal squamous cell carcinoma: downregulation of miR-143 and miR-145. World J Gastroenterol 17: 79-88.
- Ng EK, Tsang WP, Ng SS, Jin HC, Yu J, et al. (2009) MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br J Cancer 101: 699-706.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 21644
- [From(publication date):
November-2013 - Jul 03, 2025] - Breakdown by view type
- HTML page views : 16961
- PDF downloads : 4683