Research Article Open Access
Tracking the Origin of a Rabbit Haemorrhagic Virus 2 Outbreak in a Wild Rabbit Breeding Centre in Portugal; Epidemiological and Genetic Investigation
Carina L Carvalho1, Joana Rodeia2, Sandra Branco1, Madalena Monteiro3, Elsa Leclerc Duarte1, Pedro Melo4, Patricia Tavares Santos5, Ana Margarida Henriques6, Silvia Santos Barros6, Fernanda Ramos6, Teresa Fagulha6, Miguel Fevereiro6, and Margarida D Duarte6*1Department of Veterinary Medicine, Institute of Agricultural and Environmental Sciences Mediterranean, Institute for Research and Advanced Training, University of Evora, Core Mitra, 7000 Evora, Portugal
2Enterprise Development and Infrastructure Alqueva SA, Zeca Afonso Street, # 2, 7800-522 Beja, Portugal
3National Institute for Agricultural and Veterinary Research (INIAV), Pathology Laboratory, Av. Of the Republic, Quinta do Pombal, 2780-157 Oeiras, Portugal
4Vetnatura, Promenade Low of Palma, 8,7°C, 1600-176 Lisbon, Portugal
5Food and Veterinary Directorate General, Campo Grande 50, 1700-093 Lisboa, Portugal
6National Institute for Agricultural and Veterinary Research (INIAV), Virology Laboratory, Av. Of the Republic, Quinta do Pombal, 2780-157 Oeiras, Portugal
- *Corresponding Author:
- Margarida D Duarte
National Institute for Agricultural and Veterinary Research (INIAV)
Virology Laboratory, Av. Of the Republic, Quinta do Pombal, 2780-157 Oeiras, Portugal
Tel: +351 214 767 490
Fax: +351 214403666
E-mail: margarida.duarte@iniav.pt
Received date: September 20, 2016; Accepted date: October 27, 2016; Published date: November 07, 2016
Citation: Carvalho CL, Rodeia J, Branco S, Monteiro M, Duarte EL, et al. (2016) Tracking the Origin of a Rabbit Haemorrhagic Virus 2 Outbreak in a Wild Rabbit Breeding Centre in Portugal; Epidemiological and Genetic Investigation. J Emerg Infect Dis 1:114. doi:10.4172/2472-4998.1000114
Copyright: © 2016 Carvalho CL, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Infectious Disease and Pathology
Abstract
As key prey, the wild rabbit downsize constitutes a major drawback on the endangered Iberian lynx (Lynx pardinus) re-introduction in the Iberia. Several captive breeding units mostly located in Alentejo, endeavour the wild rabbit repopulation of depleted areas assigned for the lynx re-introduction.
Here we report an RHDV2 outbreak that occurred in early 2016 in a wild rabbit captive breeding unit located in Barrancos municipality. The estimated mortality rate between March and April 2016 was approximately 8.67%. Anatomopathologic examination was carried out for 13 victimized rabbits. Molecular characterization was based on the complete vp60 capsid gene.
The 13 rabbit carcasses investigated showed typical macroscopic RHD lesions testing positive to RHDV2- RNA. Comparison of the vp60 nucleotide sequences obtained from two specimens with others publically available disclosed similarities below 98.22% with RHDV2 strains originated in the Iberia and Azores and revealed that the two identical strains from Barrancos-2016 contain six unique single synonymous nucleotide polymorphisms.
In the phylogenetic analysis performed, the Barrancos-2016 strains clustered apart from other known strains, meaning they may represent new evolutionary RHDV2 lineages. No clear epidemiological link could be traced for this outbreak where the mortalities were lower compared with previous years. Yet, network analysis suggested a possible connection between the missing intermediates from which the strains from Barrancos 2013, 2014 and 2016 have derived. It is therefore possible that RHDV2 has circulated endemically in the region since 2012, with periodic epizootic occurrences.
Still, six years after its emergence in wild rabbits, RHDV2 continues to pose difficulties to the establishment of natural wild rabbit populations that are crucial for the self-sustainability of the local ecosystems.
Keywords
Rabbit haemorrhagic disease; RHDV2; Outbreak; Phylogeny; Captive wild rabbits; Iberian lynx; Conservation
Introduction
Rabbit haemorrhagic disease virus 2 (RHDV2) is a small, fastevolving RNA virus belonging to the Caliciviridae family [1].
It is known that RNA viruses’ adaptability is the result of their extreme mutation frequency, as many genomic variants are created at a high generation rate [2]. Among them, non-synonymous mutations at one or more sites may alter the RNA virus phenotypes and virulence [3,4]. Indeed in RHDV infections, like for other viruses, antigenic drift (minor changes in surface generated through point mutations) render hosts susceptible if at least a portion of the population has not been previously exposed to the new antigen(s) [5].
The mean evolution rate for RHDV based on the analysis of complete capsid gene sequences was estimated in 5.48 to 7.7 × 10-4 substitutions/site/year [6,7]. It has been suggested that the emergence of virulence occurred in the first quarter of the 20th century in a different host until it jumped to Oryctolagus spp. in China, where the disease was first reported in 1984 [7-9]. A similar process of lagovirus species jump was proposed for the emergence of RHDV2, although its putative host was not yet identified [8].
Although genetically related with RHDV, RHDV2 emergence in 2012 in Portugal had a tremendous impact in wild rabbit populations and was a major drawback for the re-introduction of the captive-bred Iberian lynx (Lynx pardinus), the most endangered wild felid in the world.
Wild rabbits are the main component of Iberian lynx diet and a keystone species on the equilibrium of the Iberian ecosystem [10]. Endemic to the Iberian Peninsula [11], the Iberian lynx population size dramatically decreased during the 20th century [12]. In 2002 was declared ‘critically endangered’ (CR) by the International Union for the Conservation of Nature (IUCN) [13,14]. Major efforts (Action Plan for the Conservation of the Iberian Lynx in Portugal; Iberian Lynx exsitu Conservation Programme; projects under the European LIFE Programme [Lince Moura/Barrancos - LIFE06 NAT/P/000191; Habitat Lince Abutre - LIFE08 NAT/P/000227; Iberlince - LIFE10 NAT/ES/000570], among others) have been made in the last decade to reverse the decline of this species. As a result, in 2015, the Iberian lynx species status was shifted to “Endangered” (EN), due to the increase of the populations in Spain. However, the RHDV2 emergence in Portugal in 2012 almost decimated the entire wild rabbit population in particular niches of mainland [15,16] and Azores [17]. A comparable scenario was also observed in Spain [18]. Although both Oryctolagus cuniculus subspecies are equally affected by RHDV2 [15,18], the susceptibility of the subspecies O. cuniculus algirus acquires special importance considering its restricted distribution to southwest [15] and the dramatic implications of RHDV2 outbreaks on its frail conservation status [16,18]. O. cuniculus algirus is a key prey species for several carnivores, including the Iberian Lynx and the Iberian imperial eagle (Aquila adalberti), both emblematic and endangered species in Portugal, and a population downsize may lead to a series of major ecological and economic problems [19].
To counteract the effects of rabbit haemorrhagic disease virus 2 (RHDV2) on the decrease of wild rabbit populations in Portugal, several measures were implemented among which restrictive hunting regulations and captive breeding of wild rabbits for subsequent return to wildlife. Rabbits restocking efforts have been recently reinforced to repopulate depleted areas in the sequence of RHDV2 outbreaks as a measure to fasten self-sustainable population establishment. Wild rabbits are breed in captivity units and set free in areas assigned either for cinegetic activity or for the Iberian lynx re-introduction. For the latter purpose, most units are located in Moura and Barrancos municipalities.
Since its first detection in Portugal in 2012 [15], RHD has been observed in some of those wild rabbit breeding units. However, to date, none of these past outbreaks were characterized and publically reported.
Here, we describe an RHDV2 outbreak in a wild rabbit captive breeding unit, built in 2004 for the wild rabbit repopulation of the Noudar Nature Park, one of the possible sites for future conservation translocations of the Iberian lynx. Phylogenetic analyses were carried out to assess the variability and putative relation of two RHDV2 Barrancos-2016 strains with other strains obtained previously in the unit, the first time in late December 2012, as well as with strains from the south, envisaging to unravel possible links between different outbreaks.
Materials and Methods
Samples and inquiry
A total of 13 victimised rabbits collected between March and April 2016, were investigated at Instituto Nacional de Investigacao Agraria e Veterinaria (INIAV). An inquiry was carried out to gather epidemiologic information on the wild rabbit captive breeding unit regarding its exact location and surroundings, extension of the parks, road accesses and animal imports (dates and animal number).
Estimation of density and mortality in 2016
Direct counts are difficult since these animals spend most of the day hidden in their warrens, making indirect counting methods preferable. Rabbits’ abundance was therefore estimated based on the intake of commercial dry rabbit food, supplied weekly to each park, assuming it was the main food source and that the average consume per rabbit per week is about 1 kg.
Mortality rates were estimated comparing the animal density inferences, calculated as described above, before and after disease outbreaks. In the unit, enumeration methods [20] (collection of cadavers) are used quite rarely for mortality estimations since they can contribute to bias results and underestimate mortality numbers as a consequence of pre-emergence death (before litters’ emerge from the nest) and predation [21].
Anatomopathologic examination
Necropsies were carried out at the University of Evora. Liver and lung samples were collected and sent to INIAV for histopathological examination. Samples were fixed in 10% buffered formalin and embedded in paraffin by standard procedures. Five micrometer-thick sections were stained with haematoxylin and eosin (H & E) prior light microscopy examination.
Virological examination
Liver and lungs samples were homogenized with phosphate buffered saline (PBS) and clarified at 3,000 g for 5 min. DNA and RNA were extracted from 200 μl of the clarified supernatant in a BioSprint 96 nucleic acid extractor (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. RHDV2-RNA was assessed by RT-qPCR [22] using the One Step RT-PCR kit (Qiagen, Hilden, Germany). Screening for classical RHDV strains was performed by conventional RT-PCR followed by sequencing analysis of the amplicons obtained with primers RC-9 and RC-10 [23], also using the One Step RT-PCR kit (Qiagen, Hilden, Germany). Myxoma virus was investigated by qPCR as described by Duarte et al. [24] with the FastStart TaqMan Probe Master Kit (Roche, Roche Diagnostics GmbH, Manheim, Germany). Cq values above 40 were considered negative.
Sequencing analysis, multiple alignments and genetic distances
Amplification of the full vp60 sequences was accomplished by two overlapping fragments obtained with primers 27F (5’-CCATGCCAGACTTGCGTCCC-3’) and 986R (5’-AACCATCTGGAGCAATTTGGG-3’) and with primers 717F (5’-CGCAGATCTCCTCACAACCC-3’) and RC10R [23].
Sequencing was carried out using a BigDye™ Terminator cycle sequencing kit (Applied Biosystems, Foster City, CA, USA), and the nucleotide sequences determined on an automated 3130 Genetic Analyzer system (Applied Biosystems, Foster City, CA, USA). The complete vp60 sequences obtained from 2 animals (strains Barrancos1/PT16 and Barrancos2/PT16) were submitted to GenBank and attributed the accession numbers KX132812 and KX132813. Nucleotide alignments were performed with Clustal Omega [25].
The percent nucleotide variability among strains based on the vp60 gene (Table 2) and the genetic divergence between populations, inferred by the nucleotide differences and substitutions between viral populations, was calculated using the DnaSP software (Version 5.10.01) [26], in order to further explore the genetic relationships among strains. Strains from the same geographic origin and year of collection were included in the same population.
Phylogenetic analysis
The phylogenetic inference was performed by Bayesian analysis. Multiple alignments of the nucleotide sequences were generated by CLUSTAL Omega [25]. The appropriated substitution model was determined resourcing to R software (R Development Core Team, 2009). The GTR model [27] with gamma-distributed rate variation across sites showed the lowest BIC and AICc values and was subsequently used to infer phylogenetic relationships using ML analysis. Robustness of the tree nodes was assessed by bootstrapping 1000 times.
For the Bayesian the CLUSTAL Omega results were converted to the NEXUS format using Mesquite software [28]. The phylogenetic tree was obtained with a Bayesian inference of phylogeny throughout the MrBayes version 3.1.2 software that uses the Markov chain Monte Carlo simulation technique to approximate the posterior probabilities of trees [29,30]. MrBayes analysis was performed using the GTR model (nst=6) with gamma-shaped rate variation with a proportion of invariable sites (rates=invgamma). The analysis was run for 106 generations (ngen=106) with four chains of temperature (nchains=4), and each chain was sampled every 10th generations (samplefreq=10).
The graphical representation and edition of the phylogenetic trees were performed with FigTree v1.3.1.
The Network software (version 5.0) was used to reconstruct a network using haplotypes (vp60 gene) by median joining [31].
Results
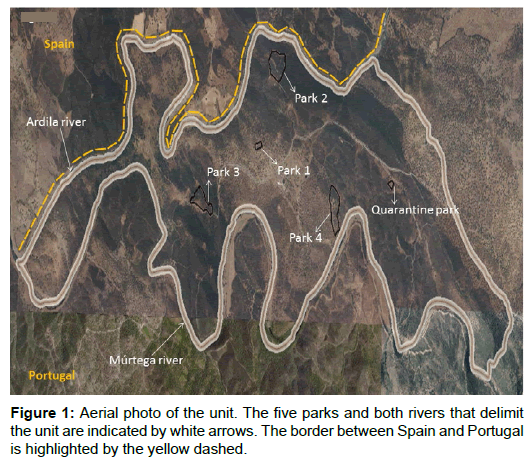
Data collected from the inquiry revealed that the unit affected by RHDV2, located in Barrancos municipality near the Spanish border, encompasses one quarantine and four reproduction parks. The five parks are sited in holm oak woodlands, only accessible by sand-clay roads, and lay on a 999 hectare extension area delimited by two riversides: southeast and within the national territory by the Múrtega River, and northwest by the Ardila River, in the Spanish border (Figure 1). Parks 1 (P1) and 3 (P3) are located closer to a busy road and therefore more exposed to human activity. In contrast, parks 2 (P2) and 4 (P4) lay on a more protected area, given their proximity to the riverside and, in the P4 case, also to a closed ore mine more distant from the roads.
The first animals (100 adults, 80 females and 20 males from subspecies O. cuniculus algirus) were introduced in 2004 (by the time of P1 construction) and originated from captive-bred wild populations from Montemor-o-Novo, located 136 Km northwest. Introduction of additional O. cuniculus algirus rabbits from other breeding parks was performed regularly (Table 1). From the second half of 2013 afterwards, following the first RHDV2 outbreak a higher number of foreign bred rabbits, mostly originated from the Spanish Granada Province, were introduced into the parks, (Table 1). The last introduction took place in 2015. The actual animal density is estimated in approximately 120 animals per hectare (≈1500 rabbits over a 12.5 hectare area). Rabbits are vaccinated against myxoma virus/ classical RHDV and identified with a tattoo or ear tag before release. No other prophylactic measures besides a coccidiostatic administration through the feed, are carried out.
Mortality by RHDV2 had been observed in the parks since 2012 (Table 1).
| Year | Causalities | |||||
|---|---|---|---|---|---|---|
| No of Parks in the unit | Translocated animals | No. of animalsc | ||||
| No. | Origin | No. of estimated casualties | Estimated mortality rate | |||
| 2009 | 2 | 55 | PT mainland | unknownc | unknownc | unknownc |
| 2010 | 2 | 173 | PT mainland | unknownc | unknownc | unknownc |
| 2011 | 2 | 121 | PT mainland | unknownc | unknownc | unknownc |
| 2012 (Dec)/2013 (Jan)a | 2 | 0 | - | 501 | 201 | 40.50% |
| 2013 (second half) | 3 | 428 | Spain and PTmainland | unknownc | - | - |
| 2014 | 4 | 664 | Spain | unknownc | 60 | 10.70% |
| 2015 | 5 | 500b | Spain | unknownc | 208 | 41.60% |
| 2016 (Jan-April) | 5 | 0 | - | 1500 | 130 | 8.67% |
| a -first outbreak of RHDV2in the unit. b-approximated number. . c-no records available | ||||||
Table 1: Information on the number and origin of animals introduced into the unit between 2009 and 2015, estimated densities and registered casualties.
In 2016, the first RHDV2 fatalities occurred in March, affecting rabbits of all ages from P1, P2 and P3. No casualties were registered in P4. Until April 2016, the mortality rate was estimated at approximately 8.67% (130/1500). No more casualties were registered from April onwards.
A total of 13 animals, corresponding to approximately 10% of the estimated casualties, were necropsied and epistaxis was observed in all animals. Haemorrhagic tracheitis, hepatic congestion and friable liver were also observed. In two animals, histopathology revealed congestion of the lungs and necrotic lesions in the liver.
All liver samples (n=13) were positive to RHDV2 by RT-qPCR [22] and negative to myxoma virus by qPCR [24]. Sequencing analysis of vp60 386 bp long fragments ruled out the presence of classical RHDV strains.
The complete vp60 nucleotide sequences (1740 bp long) obtained from 2 animals (KX132812 and KX132813) showed to be identical (100% similarity). Comparison with complete vp60 sequences publically available unrevealed the presence of six unique single synonymous nucleotide polymorphisms (SNPs) at positions 618, 940, 1020, 1077, 1440 and 1584. Blast analysis (14th June, 2016) disclosed a similarity of about 98% with strain RHDV-N11 from Spain (KM878681) as well as with several strains from across Portugal mainland and Azores.
The 2016 strains where compared with four strains from Barrancos obtained in previous years from dead wild rabbits collected in the same unit, two in January 2013 (KF442963 and KF442964) and other two in February 2014 (KM115675 and KM115676), as well as with two strains from Mértola (KM115712 and KM115713), a municipality located 81.22 km southwest from the unit, obtained in January 2014. The percent of nucleotide similarity among strains based on the vp60 gene (Table 2) and the nucleotide differences and substitutions between viral populations showed concordant values (Table 3). Besides the six SNPs already mentioned above, the Barrancos-2016 strains differ from the Barrancos-2013 and from the Barrancos-2014 strains in 26 and 37 additional residues, accounting for a total of 32 and 43 mutations, respectively (Table 3). Interestingly, the Barrancos-2016 strains showed a slightly higher nucleotide similarity and lower nucleotide differences with the Barrancos-2013 strains than with Barrancos-2014 (Tables 2 and 3). Not surprising was the lower nucleotide variability found between the Barrancos-2013 and Barrancos-2014 strains (Table 3), involving 27 nucleotide variations. Regarding strains obtained in the same year in the Barrancos municipality, a similarity of 99.94% and 100% was observed. Among the oldest (2013 and 2014) and the 2016 strains, respectively (Table 2). No elations could be taken regarding the cumulative nucleotide variation of RHDV2 since the collection of the biological materials in each year occurred always within a short period of one month.
| Barrancos | Mértola | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2016 | 2014 | 2013 | 2014 | |||||||
| Geographic origin | Year of collection | Strain name | Barrancos1PT16 | Barrancos2PT16 | CBBarrancos14-1 | CBBarrancos14-2 | 7-13_Barrancos | 10A-13_Barrancos | CBMert14-1 | CBMert14-2 |
| AC number | KX132812 | KX132813 | KM115675 | KM115676 | KF442963 | KF442964 | KM115712 | KM115713 | ||
| Barrancos | 2016 | KX132812 | - | 100 | 97.53 | 97.59 | 98.16 | 98.22 | 97.93 | 97.93 |
| KX132813 | - | 97.53 | 97.59 | 98.16 | 98.22 | 97.93 | 97.93 | |||
| 2014 | KM115675 | - | 99.94 | 98.45 | 98.51 | 97.99 | 97.99 | |||
| KM115676 | - | 98.51 | 98.57 | 98.05 | 98.05 | |||||
| 2013 | KF442963 | - | 99.94 | 98.74 | 98.74 | |||||
| KF442964 | - | 98.79 | 98.79 | |||||||
| Mértola | 2014 | KM115712 | - | 100 | ||||||
| KM115713 | - | |||||||||
| Grey indicates the range of similarities (from lighter to darker: 97.53-97.99%; 98.45-98.79%; 99.94%;100%) | ||||||||||
Table 2: Percent similarity between nucleotide sequences of vp60 gene from RHDV2 strains originated in Barrancos and Mértola municipalities.
| Total No. of mutations | No. of fixed differences | k | Average no. of nt differences between populations | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Populationsa compared | Dxy | Da | Da(JC)±SD | ||||||
| Barrancos 2016 | Vs. | Barrancos 2014 | 43 | 42 | 28,500 | 42,500 | 0,02443 | 0,02414 | 0,02454±0,01521 |
| Barrancos 2016 | Vs. | Barrancos 2013 | 32 | 31 | 21,167 | 31,500 | 0,01810 | 0,01782 | 0,01804±0,01123 |
| Barrancos 2016 | Vs. | Mértola 2014 | 37 | 35 | 24,333 | 36,000 | 0,02069 | 0,02011 | 0,02041±0,01285 |
| Barrancos 2014 | Vs. | Barrancos 2013 | 27 | 25 | 17,667 | 26,000 | 0,01494 | 0,01437 | 0,01452±0,00756 |
| Barrancos 2014 | Vs. | Mértola 2014 | 36 | 33 | 23,500 | 34,500 | 0,01983 | 0,01897 | 0,01923±0,01006 |
| Barrancos 2013 | Vs. | Mértola 2014 | 23 | 20 | 14,833 | 21,500 | 0,01236 | 0,01149 | 0,01160±0,00624 |
Table 3: Nucleotide differences and substitutions between the populations (defined as strains originated from the same municipality) considered in this study.
When comparing strains from the two municipalities, the Barrancos-2016 sequences (KX132812 and KX132813) showed 97.93% of similarity with the Mértola-2014 strains (Table 2), from which they differed in 31 nucleotides (Table 3).
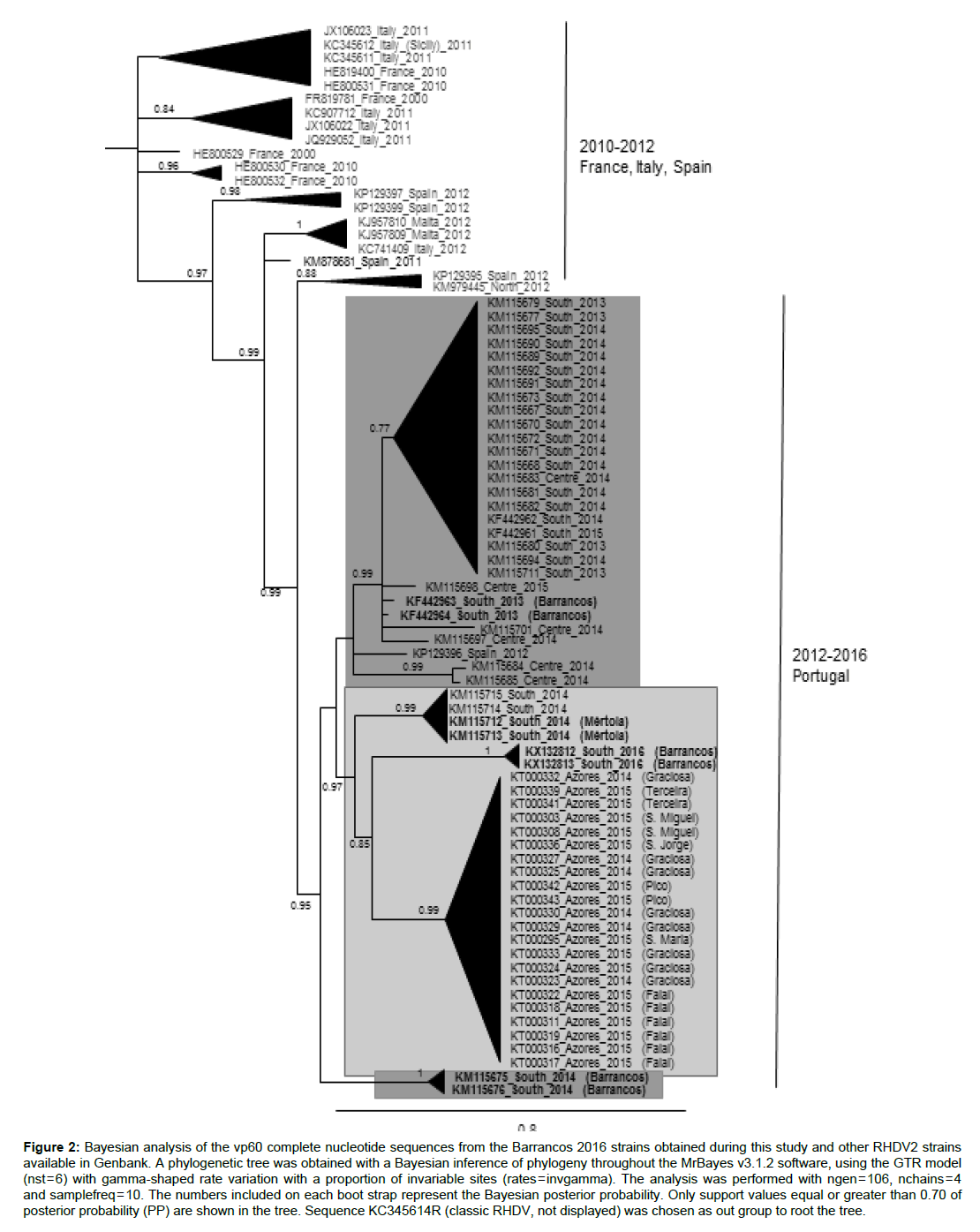
The Bayesian tree constructed with the RHDV2 sequences presently available in public databases (May 2016) revealed that the Barrancos-2016 strains are more related with the Azorean strains (Figure 2) than with any other strain. The common branch for these two groups is supported by a high posterior probability of 0.94. While the position of the strains from Barrancos-2013 and Barrancos-2014 clearly differs from Barrancos-2016, their exact relationships with other strains from mainland South remains unresolved in the cladogram (Figure 2).
Figure 2: Bayesian analysis of the vp60 complete nucleotide sequences from the Barrancos 2016 strains obtained during this study and other RHDV2 strains available in Genbank. A phylogenetic tree was obtained with a Bayesian inference of phylogeny throughout the MrBayes v3.1.2 software, using the GTR model (nst = 6) with gamma-shaped rate variation with a proportion of invariable sites (rates = invgamma). The analysis was performed with ngen = 106, nchains = 4 and samplefreq = 10. The numbers included on each boot strap represent the Bayesian posterior probability. Only support values equal or greater than 0.70 of posterior probability (PP) are shown in the tree. Sequence KC345614R (classic RHDV, not displayed) was chosen as out group to root the tree.
The second ML tree (results not shown).presented the same typology of the Bayesian tree
When all the strains obtained in the two neighbouring municipalities (Barrancos and Mértola) are compared, the Barrancos-2016 strains appear to be more closely related to the Mértola-2014 strains (KM115712 and KM115713) (Figure 2).
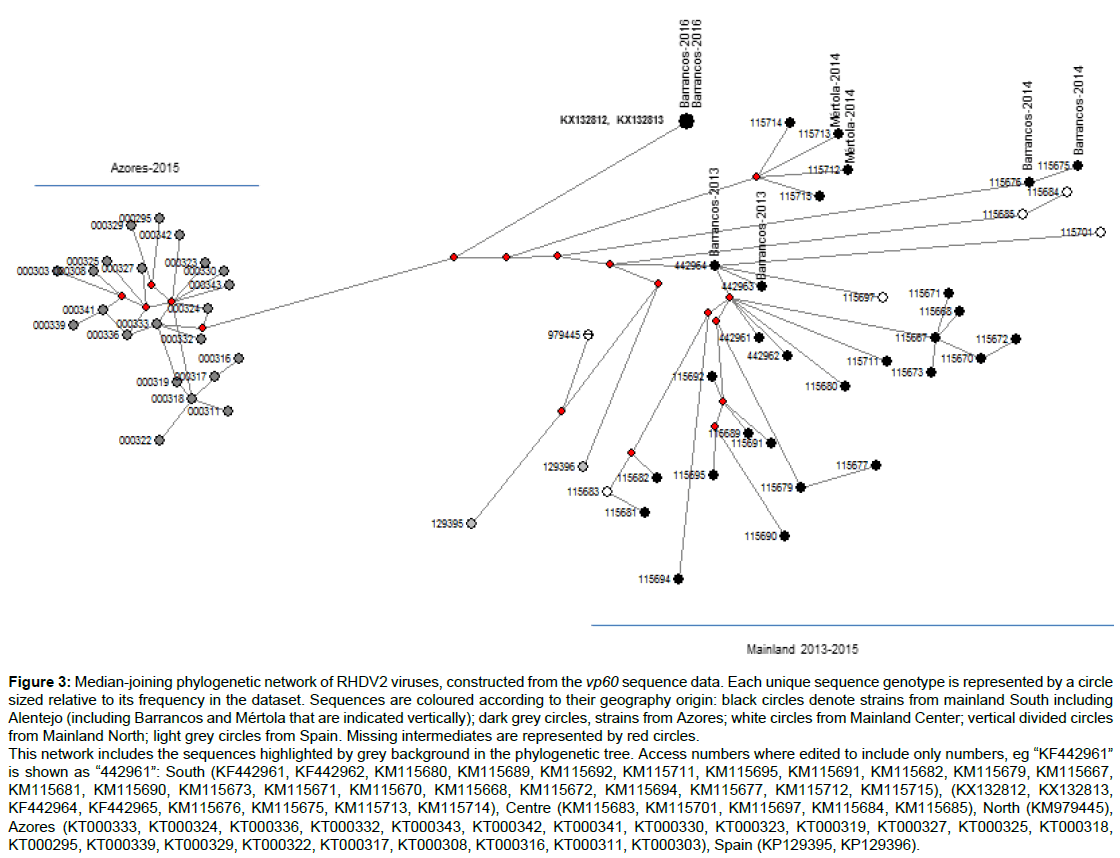
The phylogenetic network analysis (Figure 3) displayed a putative common ancestor for the RHDV2 Barrancos-2016 strains (KX132812 and KX132813) and those from Azores (KT000295, KT000303, KT000308, KT000311, KT000316-319, KT000322-25, KT000327, KT000329-330, KT000332-333, KT000336, KT000339, KT000341-343).
Figure 3: Median-joining phylogenetic network of RHDV2 viruses, constructed from the vp60 sequence data. Each unique sequence genotype is represented by a circle
sized relative to its frequency in the dataset. Sequences are coloured according to their geography origin: black circles denote strains from mainland South including
Alentejo (including Barrancos and Mértola that are indicated vertically); dark grey circles, strains from Azores; white circles from Mainland Center; vertical divided circles
from Mainland North; light grey circles from Spain. Missing intermediates are represented by red circles.
This network includes the sequences highlighted by grey background in the phylogenetic tree. Access numbers where edited to include only numbers, eg “KF442961”
is shown as “442961”: South (KF442961, KF442962, KM115680, KM115689, KM115692, KM115711, KM115695, KM115691, KM115682, KM115679, KM115667,
KM115681, KM115690, KM115673, KM115671, KM115670, KM115668, KM115672, KM115694, KM115677, KM115712, KM115715), (KX132812, KX132813,
KF442964, KF442965, KM115676, KM115675, KM115713, KM115714), Centre (KM115683, KM115701, KM115697, KM115684, KM115685), North (KM979445),
Azores (KT000333, KT000324, KT000336, KT000332, KT000343, KT000342, KT000341, KT000330, KT000323, KT000319, KT000327, KT000325, KT000318,
KT000295, KT000339, KT000329, KT000322, KT000317, KT000308, KT000316, KT000311, KT000303), Spain (KP129395, KP129396).
Also, the Barrancos-2013 strain KF442964 seems to represent a putative ancestor of KF442963 as well as for a number of strains that circulated in the South from 2013 to 2015 and Centre at least in 2014.
The analysis suggests a lineage for putative missing intermediates from which the Barrancos and Mértola strains may have arisen independently, with Mértola-2014 strains (KM115712 and KM115713) exhibiting an additional missing intermediate. Likewise, the network depicted an ancestral relation between the Barrancos-2014 strains (KM115675 and KM115676). Moreover, the two strains from Spain are more closely related with a strain from the North of Portugal mainland (KM979445), obtained from a wild rabbit in 2012 when the virus was first reported in the country [15].
Discussion and Conclusions
Immediately before release, the wild rabbits raised in the unit are vaccinated against myxoma virus and classical RHDV. However, RHDV2 vaccination, a growing practice in the domestic rabbit industry, is not practiced in this unit. Vaccination of captive wild rabbits against RHDV2 may provide temporary protection against infection and acquired immunity may increase survival of vaccinated rabbits if contact with the field strains occurs seven to 10 days after vaccination up to one year [32]. Vaccination is crucial for the establishment of herd immunity, although it can also be used as an effective post-exposure tool, in particular situations, for disease control according to the OIE [32], the difficult feasibility of vaccine administration prior return to the wild, and the limited immunity induced by a single boost raises doubts about its contribution to a faster reestablishment of natural wild rabbit populations. Since RHDV2 vaccines are not freely available on the market, their use requires special licenses from the Veterinarian Authorities. Vaccination of wild captive rabbits against RHDV2 was exceptionally authorised by Portuguese National Authority for Animal Health (DGAV) in a few other wild rabbit breeding units.
For the revival of wild populations, the development of natural immunity against RHDV2 is crucial. Many factors will affect infection outcome at the population level, such as strain virulence [9,33], herd immunity due to previous infections [4,34] population density, season of the year [7,35], anthropogenic changes in the environment [4], landscape [9] and viral transmission between populations [4,9,36].
The geographic features of the unit, circumscribed by rivers, may offer a natural barrier to animal movement which is expected to slow down virus dissemination within the parks, similarly to what was described for RHDV [7,37], giving the virus more time to evolve [9]. This might explain the genetic diversity found between the strains from this unit obtained across this 4-years period. Also, the rapid transmission of a pathogen will affect the local spread of the disease reducing the number of susceptible individuals [4]. Hence, RHDV2 moderate virulence compared to RHDV, constitutes a selective advantage of this virus [9], as it was demonstrated that moderately virulent RHDV-related strains can invade wild rabbit populations with greater efficiency than highly virulent or non-pathogenic strains [9,33].
Interestingly, despite all the other parks in the unit were affected by RHDV2, there were no casualties recorded in P4. Since the virus first detection in the unit in 2012, animals from P4 seem to be passing unharmed. Although the factors behind the lower incidence of infection in P4 are not fully clarified, it is likely that its geographic isolation may have protected rabbits more efficiently from infection. However, the higher levels of humidity near the riverside probably also favour mosquitoes, which are often implicated in disease transmission as mechanic vectors for RHDV [38], therefore puzzling this uneven occurrence of disease.
Also, an artificial boundary by electric fencing delimits each park adding up to the protection from predators given by several natural shelters located within the parks. Yet, the involving forestland provides suitable habitats for many free-ranging species including wild rabbits and small predators and scavengers such as badgers (Meles meles), mongooses (Herpestes ichcneumon), weasels (Mustela nivalis), foxes (Vulpes vulpes), stone martens (Martes foina), as well as birds of prey, often seen in the area, whose proximity with the parks may favour the direct or indirect contact with captive wild rabbits. Some predators and scavengers species can remove small animal carcasses from the site of death to other locations [39] and, concurring with that, a few specimens collected during this outbreak were damaged, probably due to scavenging. In fact, consumption of RHDV-victimized rabbits may be at the origin of the RHDV antibodies detected in red foxes and other predators and scavengers living in sympatry with affected rabbit populations [40-42]. These serological evidences pointed to the possibility of other species, apart from rabbits, being involved in the epidemiology and persistence of the disease [43].
Given the complex eco-epidemiology of RHDV2 depicted above, while efficient herd immunity is not achieved, serious difficulties may be posed to the establishment of self-sustainable wild rabbits populations in Baixo-Alentejo.
The phylogenetic analysis did not allow clarifying the origin of the infection since the Barrancos-2016 strains did not group with any other strain presently known, showing however to be more closely related with strains from Azores (supported by a posterior probability of 0.85) (Figure 2). When looking to viruses originated in the same district, the Bayesian tree showed that Barrancos-2016 strains are more closely related with the Mértola-2014 strains than with other strains from Barrancos (pp of 0.99). The Barrancos-2013 strains clustered with strains from the South and Centre of Portugal mainland, while the two Barrancos-2014 strains grouped independently in a more ancestral branch, contradicting the temporal structure with regards to the position in the tree. However, these results were further clarified by the phylogenetic network analysis performed (Figure 3) where missing intermediates from which the Barrancos 2013, 2014 and 2016 strains may have evolved, present a chronological relative correct position. Based on the phylogenetic analyses, the Barrancos 2016 could represent an independent introduction of RHDV2 from unknown origin. It is however worthy of mention that the unit is located in a preserved area were hunting activity is not allowed, therefore movement of hunters, which could be implicated in the virus dissemination from other locations, did not account for the occurrence of the outbreak.
Despite disease reports in the last five years suggest that RHDV2 may have circulated endemically in the region with epizootic occurrences, mortality was not observed in the immediate surrounding areas of the parks. Nonetheless, the possible involvement of the local wild rabbit population in the outbreak couldn’t be proven or excluded, since the outside area was not systematically investigated prior the outbreak, a limitation of this study. The new virulent Barrancos-2016 strains could have evolved from previous circulating strains through cumulative mutations. The phylogenetic similarity with other strains isolated from Portugal, even though with no apparent geographical or epidemiological link, is more in favour of a common infection source rather than a less probable convergent evolution.
Finally, regarding rabbit translocations, the last introduction of animals in the park took place in January 2015, 14 months before the 2016 fatalities. It is unlikely that the virus was introduced from the Spain park supplier, where no records of RHDV2 infection were reported. Furthermore, during the at least 15 day quarantine carried out before translocation, no clinical signs of disease were observed.
The mortality rate estimations can contain some bias, a limitation inherent to the used methodology and related to the fact that in P2 and P3, in contrast with P1, rabbits have access to some seasonal natural food source, like acorn and grass, being also more vulnerable to predation, particularly by birds of prey. Nevertheless, the mortality rate estimated in 2016 was comparably lower than that observed in former years, namely in the outbreak between December 2012 and January 2013 (≈200 victimized animals) when the virus was first detected in the unit. At the time, the centre encompassed two parks with 200 and 300 animals, were a mortality rate of 42% and 39% (average 40.5%) was registered. This lower mortality rate compared with other reported outbreaks may be the reflex of a gradual RHDV2 immunity development in wild rabbit populations and its passive transmission to the offspring, and a promising sign towards the establishment of hostvirus equilibrium.
References
- Dalton KP, Abrantes J, Lopes AM, Nicieza I, Alvarez AL, et al. (2015) Complete genome sequence of two rabbit hemorrhagic disease virus variant b isolates detected on the Iberian Peninsula. Arch Virol 160: 877-881.
- Holmes EC (2004) The phylogeography of human viruses. Mol Ecol 13: 745-756.
- Domingo E, Martínez-Salas E, Sobrino F, de la Torre JC, Portela A, et al. (1985) The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance--a review. Gene 40: 1-8.
- Boots M, Hudson PJ, Sasaki A (2004) Large shifts in pathogen virulence relate to host population structure. Science 303: 842-844.
- Harrison LH, Jolley KA, Shutt KA, Marsh JW, O’Leary M, et al. (2006) Maryland Emerging Infections Program. Antigenic shift and increased incidence of meningococcal disease. J Infect Dis 193-1266-1274.
- Kerr PJ, Kitchen A, Holmes EC (2009) Origin and phylodynamics of rabbit hemorrhagic disease virus. J Virol 83: 12129-12138.
- Alda F, Gaitero T, Suarez M, Merchan T, Rocha G, et al. (2010) Evolutionary history and molecular epidemiology of rabbit haemorrhagic disease virus in the Iberian Peninsula and Western Europe. BMC Evol Biol 10: 347.
- Liu SJ, Xue HP, Pu BQ, Qian NH (1984) A new viral disease of rabbits. Animal Husbandry Vet Med 16: 253-255.
- Le Gall-Recule G, Lavazza A, Marchandeau S, Bertagnoli S, Zwingelstein F, et al. (2013) Emergence of a new lagovirus related to Rabbit Haemorrhagic Disease Virus. Vet Res 44: 81.
- Delibes-Mateos M, Delibes M, Ferreras P, Villafuerte R (2008) Key role of European rabbits in the conservation of the Western Mediterranean basin hotspot. Conserv Biol 22: 1106-1117.
- Ferrer M, Negro JJ (2004) The Near Extinction of Two Large European Predators: Super Specialists Pay a Price. Conserv Biol 18: 344-349.
- Rodríguez A, Delibes M (1992) Current range and status of the Iberian lynx Felis pardina Temminck, 1824 in Spain. Biol Conserv 61: 189-196.
- IUCN (2003) IUCN Red List of Threatened Species.
- Simon MA, Gil-Sanchez JM, Ruiz G, Garrote G, McCain EB, et al. (2012) Reverse of the decline of the endangered Iberian lynx. Conserv Biol 26: 731-736.
- Abrantes J, Lopes AM, Dalton KP, Melo P, Correia JJ, et al. (2013) New variant of rabbit hemorrhagic disease virus, Portugal, 2012-2013. Emerg Infect Dis 19: 1900-1902.
- Lopes AM, Correia J, Abrantes J, Melo P, Ramada M, et al. (2015) Is the new variant RHDV replacing genogroup 1 in Portuguese wild rabbit populations? Viruses 7: 27-36.
- Duarte M, Henriques M, Barros SC, Fagulha T, Ramos F, et al. (2015) Detection of RHDV variant 2 in the Azores. Vet Rec 176: 130.
- Delibes-Mateos M, Ferreira C, Carro F, Escudero MA, Gortazar C (2014) Ecosystem effects of variant rabbit hemorrhagic disease virus, Iberian Peninsula. Emerg Infect Dis 20: 2166-2168.
- Delibes-Mateos M, Ferreras P, Villafuerte R (2007) Rabbit populations and game management: the situation after 15 years of rabbit haemorrhagic disease in central-southern Spain. Biodivers Conserv 17: 559-574.
- Marchandeau S, Aubineau J, Berger F (2006) Abundance indices: reliability testing is crucial-a field case of wild rabbit Oryctolagus cuniculus. Wildlife Biol.
- Norbury G (2001) Advances in New Zealand mammalogy 1990–2000: lagomorphs. J R Soc New Zeal.
- Duarte MD, Carvalho CL, Barros SC, Henriques AM, Ramos F, et al. (2015) A real time Taqman RT-PCR for the detection of rabbit hemorrhagic disease virus 2 (RHDV2). J Virol Methods 219: 90-95.
- Tham KM, Barnes SM, Hunter SN (1999) Polymerase chain reaction amplification and gene sequence analysis of a calicivirus from a feral rabbit. Virus Genes 18: 235-242.
- Duarte MD, Barros SC, Henriques AM, Fagulha MT, Ramos F, et al. (2014) Development and validation of a real time PCR for the detection of myxoma virus based on the diploid gene M000.5L/R. J Virol Methods 196: 219-224.
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539.
- Librado P, Rosas (2009) DNA Sp v.5 A software for comprehensive analysis of DNA polymorphism data. Bioinformatis 25: 1451-1452
- Tavare S (1986) Some probabilistic and statistical problems in the anal- ysis of DNA sequences. In: Miura RM (Ed). Lectures on math- ematics in the life sciences. Volume 17. Providence (RI):American Mathematical Society. 57-86.
- Maddison WP, Maddison DR (2009) Mesquite: a modular system for evolutionary analysis.
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP (2001) Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294: 2310-2314.
- Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572-1574.
- Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16: 37-48.
- OIE Terrestrial manual (2016) Rabbit haemorrhagic disease.
- Fouchet D, Le Pendu J, Guitton JS, Guiserix M, Marchandeau S, et al. (2009) Evolution of microparasites in spatially and genetically structured host populations: the example of RHDV infecting rabbits. J Theor Biol 257: 212-227.
- White PJ, Norman RA, Hudson PJ (2002) Epidemiological consequences of a pathogen having both virulent and avirulent modes of transmission: the case of rabbit haemorrhagic disease virus. Epidemiol Infect 129: 665-677.
- Cooke BD (2002) Rabbit haemorrhagic disease: field epidemiology and the management of wild rabbit populations. Rev Sci Tech 21: 347-358.
- Calvete C (2006) Modeling the effect of population dynamics on the impact of rabbit hemorrhagic disease. Conserv Biol 20: 1232-1241.
- Muller A, Freitas J, Silva E, Le Gall-Recule G, Zwingelstein F, et al. (2009) Evolution of rabbit haemorrhagic disease virus (RHDV) in the European rabbit (Oryctolagus cuniculus) from the Iberian Peninsula. Vet Microbiol 135: 368-373.
- McColl KA, Merchant JC, Hardy J, Cooke BD, Robinson A, et al. (2002) Evidence for insect transmission of rabbit haemorrhagic disease virus. Epidemio Infect 129: 655-663.
- Santos SM, Carvalho F, Mira A (2011) How long do the dead survive on the road? Carcass persistence probability and implications for road-kill monitoring surveys. PLoS One 6: e25383.
- Frölich K, Klima F, Dedek J (1998) Antibodies against rabbit hemorrhagic disease virus in free-ranging red foxes from Germany. J Wildl Dis 34: 436-442.
- Leighton FA, Artois M, Capucci L, Gavier-Widen D, Morisse JP (1995) Antibody response to rabbit viral hemorrhagic disease virus in red foxes (Vulpes vulpes) consuming livers of infected rabbits (Oryctolagus cuniculus). J Wildl Dis 31: 541-544.
- Parkes JP, Heyward RP, Henning J, Motha MXJ (2004) Antibody responses to rabbit haemorrhagic disease virus in predators, scavengers, and hares in New Zealand during epidemics in sympatric rabbit populations. NZ Vet J 52: 85-89.
- Merchan T, Rocha G, Alda F, Silva E, Thompson G, et al. (2011) Detection of rabbit haemorrhagic disease virus (RHDV) in nonspecific vertebrate hosts sympatric to the European wild rabbit (Oryctolagus cuniculus). Infect Genet Evol 11: 1469-1474.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 11348
- [From(publication date):
December-2016 - Apr 16, 2025] - Breakdown by view type
- HTML page views : 10437
- PDF downloads : 911