Towards Structural Based Drug Development for Ebola Virus Disease
Received: 20-Feb-2016 / Accepted Date: 24-Feb-2016 / Published Date: 29-Feb-2016 DOI: 10.4172/2572-0406.1000e101
9487Editorial
Ebola virus disease (EVD), characterized by fatal bleeding and coagulation abnormalities, is caused by infection of Ebola viruses (EBOV) and other members of the family Filoviridae. Ebola viruses have 5 species named after the places of outbreaks: Zaire ebolavirus (ZEBOV), Bundibugyo ebolavirus (BDBV), Reston ebolavirus (RESTV), Sudan ebolavirus (SUDV), and Taï Forest ebolavirus (TAFV). Among them, the Zaire strain is the most lethal. The EBOV genome consists of a linear, non-segmented negative-stranded RNA of 19 kb in length, coding for 7 structural proteins, including nucleoprotein (NP), VP35, VP40, glycoprotein (GP), VP30, VP24, and L protein. Human infection by EBOV is possible through contact with body fluids of virus infected individuals or animals like Primates or fruit bats. There have been over 25 outbreaks since its discovery in 1976. In the most recent largest Ebola outbreak in Western Africa, 28,639 cases and 11,316 deaths were recorded by the World Health Organization (WHO, http://www.who.int/csr/disease/ebola/situationreports/ en/). There is no Food and Drug Administration (FDA) approved specific treatment for the EVD. The medical care for patients primarily relies on intensive supportive care. The use of convalescent plasma from patients who have recovered from EVD was among the first specific therapeutic approaches, and later on, human neutralizing antibodies, like ZMappTM, have been tested. However, the source of convalescent plasma is very limited and antibody production is too expensive to meet the demands for large scale applications, especially for those patients in less developed countries most at risk from Ebola outbreaks. Using small interfering RNA (siRNA) targeting Ebola Virus (EBOV) genome is another appealing therapeutic idea. A successful siRNA product, TKM-Ebola, developed by the Tekmira pharmaceuticals corp, has been tried clinically during Ebola crisis. It uses a mixture of three siRNAs to target EBOV VP24 (membrane associated protein), VP35 (polymerase complex protein) and L (RNA dependent RNA polymerase) respectively. It is effective against Ebola virus, however, the TKM-Ebola therapy was discontinued soon after phase I clinical trials due to activation of inflammatory pathways in patients.
Treatment with small chemical drugs would be ideal, as these are easily manufactured, stable, inexpensive and can be administered orally. In the absence of specific chemical drug therapies, existing FDA approved drugs have been tested for possible repurposing usage for EVD. These drugs already have good safety records from previous clinical studies and allow a faster transition into clinical trials. Among them, three chemical drugs, Brincidofovir (CMX001, a nucleotide analogue used against cytomegalovirus), Amiodarone (an antiarrhythmic drug) and Favipiravir (T-705, a pyrazine carboxamide derivative used for influenza infection) have been used for clinical trials during recent West African Ebola crisis. These chemical drugs are not specific to the Ebola virus and the therapeutic benefits have not been clearly demonstrated. Many other FDA approved drugs have been identified using in vitro anti-Ebola assays, some of which have been proven to be effective against Ebola virus in in vivo animal models. Now, the priority is to find out targets for those drugs and have the target-drug complex structures. Those repurposed drugs may be not of high potency as it is however, the structural information would be very valuable for more powerful drug development.
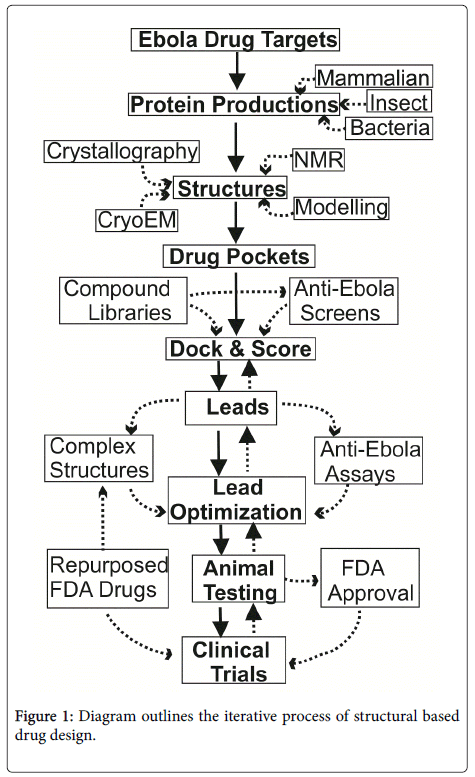
The basic procedures for structural based drug discovery is shown in Figure 1. First of all, protein targets have to be selected from the Ebola virus or its receptors. The target genes need to be cloned into protein expression systems, either for bacteria, insect-baculovirus or mammalian expression. Usually, large scale expressions are required to achieve milligram amounts of pure proteins. These proteins can be used for structure determination with one of following approaches: nuclear magnetic resonance spectroscopy (NMR, for smaller sized proteins, up to ~ 50 kd), cryo-electron microscope (cryo-EM, for larger proteins, minimum size of 120 kd) and mostly X-ray crystallography. From structures, potential drug binding pockets may be identified. With computer docking software packages, chemical compounds can be placed into the pockets. These compounds are scored and ranked based on their steric and electrostatic interactions with the pocket lining protein residues, and best compounds can be chosen to test their anti-Ebola efficacy. Then, complex structures with these promising leads are required to reveal the compound sites that can be optimized to increase potency. Many cycles of complex structure optimization may be required to identify specific and powerful compounds.
To select targets for EBOV drug development, the viral GP and its receptors, like Niemann-Pick type C1 (NPC1) may be first line candidates. Much progress has recently been made in determining the structures of EBOV GP and its receptor NPC1 [1-3].
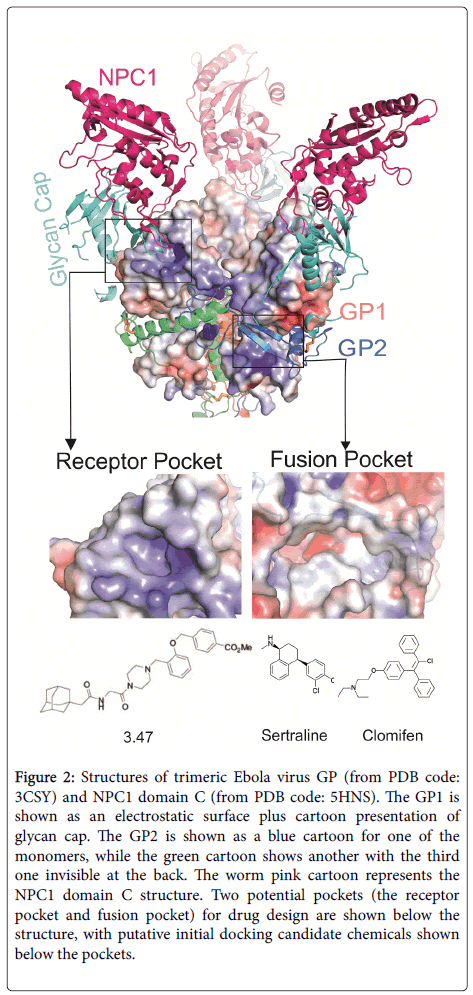
Figure 2 shows the trimeric EBOV GP structure with its receptor NPC1 luminal domain C (NPC1 DC). The EBOV trimeric glycoprotein’s spikes on the viral membrane envelope are solely responsible for host cell entry. The GP is processed into two subunits, GP1 and GP2, by proteases within the Golgi apparatus, but two subunits are still held together by a disulphide bond. The GP1 subunit is responsible for receptors binding, whereas the GP2 subunit is responsible for fusing the viral membrane to the host cell endosome membrane. The viral GP initiates cell attachment by binding to some nonspecific attachment factors, such as C-type lectins, phosphatidylserine receptors, T-cell immunoglobulin domain-1(TIM-1) and tyrosine protein kinase receptor 3 (TYRO3). After a micropinocytosis like process, the virus containing vesicles traffic through early and late endosomes, where the lower pH activates the endosomal proteases cathepsin B and / or L to trim off the glycan cap including the mucin domain of GP1 (Figure 2, cyan coloured cartoon) , and expose the receptor ligand binding domain (LBD). The NPC1 receptor binds to the GP LBD with its domain C (Figure 2, warm pink coloured cartoon). With the help of other factors, for example, the two pore calcium channel protein 2 (TPC2), the fusion of viral membrane with host cell endosome membrane is triggered. The viral genome is released into the host cell cytosol and begins replication. Chemical compounds targeting the EBOV GP or its receptors NPC1, TPC2 would disrupt the viral entry and prevent the infection. With recently determined GP and NPC1 structures, some potential drug pockets could be identified (Figure 2). In the GP1 LBD, the pocket used for NPC1 DC binding may serve as a useful potential pocket for drug design (Figure 2, receptor pocket). The pocket is covered by GP1’s own glycan cap. Interestingly, the neutralizing antibody MR78 also binds to this pocket [4]. Some chemicals like compounds 3.47 have been previously demonstrated to interfere the GP-NPC1 interaction [5]. Whether this chemical directly binds to GP or NPC1 still remain to be investigated. Another possible EBOV GP “druggable” pocket is located in GP1 and GP2 interface near the fusion loop (Figure 2, fusion pocket). Therefore, chemicals that bind to this pocket may interfere with the fusion process, and compound 7 has been predicted to bind this pocket [6].
Figure 2: Structures of trimeric Ebola virus GP (from PDB code: 3CSY) and NPC1 domain C (from PDB code: 5HNS). The GP1 is shown as an electrostatic surface plus cartoon presentation of glycan cap. The GP2 is shown as a blue cartoon for one of the monomers, while the green cartoon shows another with the third one invisible at the back. The worm pink cartoon represents the NPC1 domain C structure. Two potential pockets (the receptor pocket and fusion pocket) for drug design are shown below the structure, with putative initial docking candidate chemicals shown below the pockets.
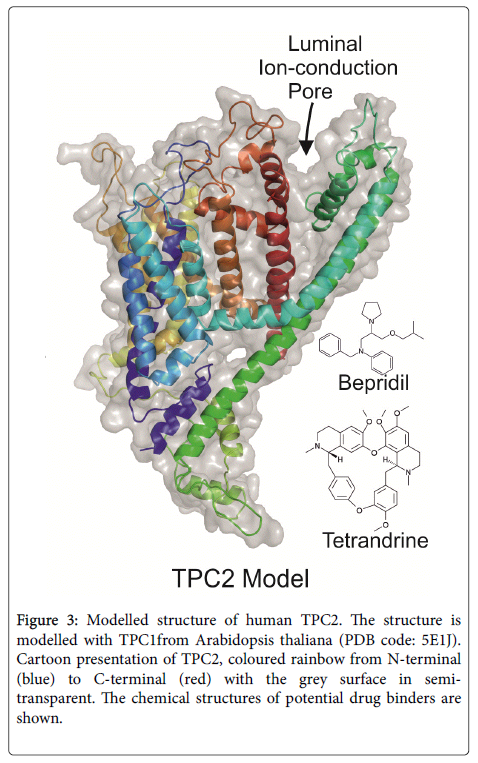
Despite many advances in computing techniques, it remains a major challenge to design a de novo compound on the basis of an unliganded pocket. Structures of a compound bound pocket would tremendously facilitate the drug design and optimization. Unfortunately, none of these long waited complex structures are available. In the last few years, many chemicals have been screened for EBOV entry inhibition with in vitro cellular assays. Among these tested chemicals, sertraline [7] and clomifen [8] (chemical structures shown in Figure 2) and some others have been shown good inhibition of Ebola infection in vitro , and even in the in vivo animal models. These chemicals may serve as priority candidates for co-crystallization with EBOV GP or NPC1. The structure for another important EBOV entry factor TPC2 calcium ion channel is not available. However, a similar structure of TPC1 from plant mouse ear cress (Arabidopsis thaliana) has recently been solved [9]. The human TPC2 structure can be modelled with the plant TPC1, as shown in Figure 3.
Figure 3: Modelled structure of human TPC2. The structure is modelled with TPC1from Arabidopsis thaliana (PDB code: 5E1J). Cartoon presentation of TPC2, coloured rainbow from N-terminal (blue) to C-terminal (red) with the grey surface in semitransparent. The chemical structures of potential drug binders are shown.
Some calcium ion channel blockers, like beprilil [7] and tetrandrine [10] (chemical structures shown in Figure 3) have been selected from anti-Ebola cellular assays. The actual human TPC2 structure in complex with some of these chemicals would greatly enhance structural based drug development.
Despite the progress in Ebola virus biochemical and structural studies, we are still in the early stages of the anti-Ebola drug development cycle. Complex structures of chemical compounds with Ebola virus proteins or viral receptors are desperately needed. With great advancements of high performance computational approaches, we are optimistic about seeing rapid progress towards drug development for EVD therapy.
References
- Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, et al. (2008) Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454: 177-182.
- Wang H, Shi Y, Song J, Qi J, Lu G, et al. (2016) Ebola viral glycoprotein bound to Its endosomal receptor Niemann-Pick C1. Cell 164: 258-268.
- Zhao Y, Ren J, Harlos K, Stuart DI (2016) Structure of glycosylated NPC1 luminal domain C reveals insights into NPC2 and Ebola virus interactions. FEBS lett.
- Hashiguchi T, Fusco ML, Bornholdt ZA, Lee JE, Flyak AI, et al. (2015) Structural basis for Marburg virus neutralization by a cross-reactive human antibody. Cell 160: 904-912.
- Cote M, Misasi J, Ren T, Bruchez A, Lee K, et al. (2011) Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477: 344-348.
- Basu A, Li B, Mills DM, Panchal RG, Cardinale SC, et al. (2011) Identification of a small-molecule entry inhibitor for filoviruses. J Virol 85: 3106-3119.
- Johansen LM, DeWald LE, Shoemaker CJ, Hoffstrom BG, Lear-Rooney CM, et al. (2015) A screen of approved drugs and molecular probes identifies therapeutics with anti-Ebola virus activity. Science translational medicine 7: 290ra89.
- Johansen LM, Brannan JM, Delos SE, Shoemaker CJ, Stossel A, et al. (2013) FDA-approved selective estrogen receptor modulators inhibit Ebola virus infection. Science translational medicine 5: 190ra79.
- Guo J, Zeng W, Chen Q, Lee C, Chen L, et al. (2 015) Structure of the voltage-gated two-pore channel TPC1 from Arabidopsis thaliana. Nature.
- Sakurai Y, Kolokoltsov AA, Chen CC, Tidwell MW, Bauta WE, et al. (2015) Ebola virus. Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science 347: 995-998.
Citation: Zhao Y (2016) Towards Structural Based Drug Development for Ebola Virus Disease. J Chem Biol Ther 1: e101. DOI: 10.4172/2572-0406.1000e101
Copyright: © 2016 Zhao Y. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Open Access Journals
Article Tools
Article Usage
- Total views: 12764
- [From(publication date): 2-2016 - Mar 31, 2025]
- Breakdown by view type
- HTML page views: 11734
- PDF downloads: 1030