The Expression of DNA Methyltransferase DNMT3a in Classical and Fibrolamellar Hepatocellular Carcinoma
Received: 06-Jun-2016 / Accepted Date: 29-Jun-2016 / Published Date: 30-Jun-2016 DOI: 10.4172/2161-0681.1000282
Abstract
The epigenetic regulation of DNA-templated processes has been studied extensively during the last 15 years. As was revealed, DNA modification such as methylation possess great impact on cell fate and can result in abnormal protein expression patterns what can lead to the induction of carcinogenesis. DNA (cytosine-5)-methyltransferases are enzymes that catalyses the transfer of methyl groups to specific CpG structures in DNA. The methylation of these sequences can lead to inappropriate gene expression such as the silencing of tumor suppressor genes in cancer cells. DNA (cytosine-5)-methyltransferase 3A (DNMT3a) gene encodes a DNA methyltransferase that is thought to function mainly in de novo methylation. In the normal liver DNMT3a is usually expressed on the medium level, as was described in literature. Hepatocellular carcinoma (HCC) still remains one of the most common cause of death among patients with cancer. Fibrolamellar hepatocellular carcinoma (FL) represents rare subtype, which affects usually young people (an onset between 20-30 years) and its etiology is poorly understood. In our study we compared the presence of DNMT3a protein between two different types of HCC-common type and fibrolamellar one. We performed immunohistochemical staining of formalin fixed paraffin embedded tissue sections obtained from 30 patients (22 HCC and 8 FL). We found that DNMT3a immunoreactivity is significantly more pronounced in the non-fibrolamellar variant of HCC than in the fibrolamellar one. The DNMT3a immunoreactivity was predominantly localized in cancer cell nuclei in a form of separate large granules spotted in proximity to heterochromatin region. The reduced presence of DNMT3a in the fibrolamellar variant of HCC may suggest that different epigenetic mechanisms are involved in development of this particular type of liver cancer. Improving our understanding of the roles of DNMT proteins in hepatocarcinogenesis can benefit in the development of epigenetic –based therapy designed for specific HCC subtype.
Keywords: DNMT3a ; Methyltransferase; Hepatocellular carcinoma; Fibrolamellar carcinoma
315078Introduction
The epigenetic regulation of DNA-templated processes, such as transcription, replication or DNA repair has been studied extensively during the last 15 years. This discovery brought a significant alteration of the so-called central dogma of molecular biology, as far as the sequence of the DNA bases is no longer considered to be the sole determining factor of gene expression.
As was revealed, DNA modification such as methylation, or histone acetylation possess great impact on cell fate and can result in abnormal protein expression patterns what can lead to the induction of carcinogenesis. Furthermore, the role of epigenetic changes in genome has been discovered in such processes as regulation of metabolism, cell differentiation, tissue regeneration, memory processes, individual difference in immunological responses to infection [1]. The spectrum of diseases, in which dysregulation of epigenetic processes occurs is equally wide: it includes, apart from cancer, autoimmunological diseases (e.g. rheumatoid arthritis, multiple sclerosis, diabetes type 1), neurodegenerative disorders, not to mention hereditary syndromes associated with parental or maternal methylation of specific loci (Silver-Russell syndrome, Beckwith-Wiedemann syndrome, Angelmann syndrome, Prader-Willi syndrome) [2].
DNA (cytosine-5)-methyltransferases are enzymes that catalyse the transfer of methyl groups to specific CpG structures in DNA. The methylation of these sequences can lead to inappropriate gene silencing, such as the silencing of tumour suppressor genes in cancer cells. The demethylation of repeated DNA sequences has also been researched in tumours. In the former group the following demethylated genes have the greatest clinical importance: HOX1 in Tcell acute lymphoblastic leukaemia, MAGE gene family in melanoma and DMR-LIT1 in esophageal cancer [3]. The loss of methylation occurs chiefly in CpG groups localised mainly in promoters, however demethylation sites in exons and introns have also been observed [3]. The repeated DNA sequences which are subject to demethylation include LINE-1 and HERV-K . The role of this process in carcinogenesis was not clearly elucidated, it is supposed that hypomethylated LTRs demonstrate higher frequency of homologous recombination and thus lead to chromosome rearrangements and genome instability [4].
On the other hand, the loss of methylation in proto-oncogenes, described as active demethylation and mediated by TET1 and TET2 proteins might augment their expression and form the initial steps in carcinogenesis [5].
The family of mammalian methyltransferases consists of five enzymes, of which three possess enzymatic activity for DNA methylation - the remaining member of the group, DNMT2 plays an important role in polypeptide synthesis through tRNA methylation, hence its alternative name: TRDMT1 (tRNA aspartic acid methyltransferase 1) [6].
DNMT1 is considered to be the chief DNA methylase participating in maintaining the current methylation status. Its activity is revealed against hemimethylated CpG islands. It means that the enzyme restores the methylation status in loci, where of the strands lost its methylation during a spontaneous process caused passive (i.e., not enzyme-mediated) demethylation. However the expression of this enzyme in carcinomatous tissue correlates poorly with total hypomethylation or hypomethylation of repeats. Moreover, the expression of DNMT1 seems to be increased, not silenced in majority of tumours [7].
DNMT3a gene encodes a DNA methyltransferase that is thought to function in de novo methylation, rather than the maintenance of existing methylated sites. Other related proteins include DNMT3b (lower enzymatic activity than DNMT3a ) and DMNT3L (a postulated regulatory role, no enzymatic activity). This distinction originates in specificity of enzymatic activity: whereas DNMT1 methylates hemimethylated sites 100 times more efficiently than non-methylated CpG islands, DNMT3a has no such predispositions and is equally active no matter what the previous methylation status was [8]. It has been reported that aberrant hypermethylation of CpG islands is implicated in carcinogenesis. Especially acute myeloid leukaemia (AML) has been associated with mutations in DNMT3a , which are an independent factor of poor prognosis [9]. Its elevated expression was observed in gastric cancer as well [10]. Its role has also been established in the development of hepatocellular carcinoma (HCC) - inhibition of DNMT3a expression by microRNA miR-200b causes decrease in tumour proliferative activity [11]. On the other hand, the expression of DNMT3a is up-regulated by coexisting hepatitis B virus (HBV) infection [12], which is one of the main risk factor for development of HCC with the significant exception of fibrolamellar variant.
Hepatocellular carcinoma still remains one of the most common cause of death among patients with cancer. Especially patients with higher stage of cancer at the moment of diagnosis have poor prognosis (5-year survival rate at 2% for IVB and 7% for IIIC and IVA). The main etiological factor of this tumour is cirrhosis, regardless of its causative agent (viral B or C hepatitis, autoimmune hepatitis, alcoholism) [13]. However, one specific subtype of HCC, fibrolamellar (FL), does not arise on the basis of cirrhotic liver and its etiology remains unknown. Its distinct pathomorphologic features (plenty of mitochondria in cells, whose groups are separated by thick collagen septa) come in line with different clinical features. This type of HCC affects usually young people (an onset between 20-30 years) without gender preponderance (as opposed to 3-8:1 M:F ratio for other types of HCC) [14]. It is therefore postulated that pathomechanism of carcinogenesis and molecular changes might also be distinct for these tumour subsets.
Objectives
The aim of the study was to assess the presence of DNMT3a protein in two different types of hepatocellular carcinoma - common type and fibrolamellar one, as well as in the normal liver both in quantitative and qualitative (intracellular localisation) terms using immunohistochemistry. Furthermore we analysed the mRNA levels of DNMT3a in HCC and in the normal liver.
Materials and Methods
Tumor samples
We performed our study on 20 common type, 5 fibrolamellar hepatocellular carcinomas and 11 non-malignant liver tissues.
Immunohistochemistry (IHC)
Formalin fixed paraffin embedded sections were subjected to peroxidase-based antigen immunodetection with the anti-DNMT3a (Atlas Antibodies, USA) antibody. Immunostaining was performed using Leica BOND Automated Immunostainer (Leica Biosystems, Germany).
Real-time PCR
In two experimental groups (normal liver and non-fibrolamellar HCC) DNMT3a expression was evaluated by RT-qPCR. RNA was isolated using Direct-zol™ RNA MiniPrep (ZymoResearch, USA). Gene expression analysis was performed using Taq Man Gene Expression Assay (Invitrogen), on AbiPrism 7500 (Applied Bioststems, USA) system. Molecular probes complementary to mRNA sequences of all the genes were purchased from Invitrogen (USA), stained by the manufacturer with FAM as a fluorophore and TAMRA as a quencher. GAPDH was selected as the reference gene. Expression levels were counted using 2-DDCt method and estimated as a fraction of reference gene expression.
Data analysis
The immunoreactivity was quantified by the assessment of the percentage of immunopositive cells counted in 5 representative high power fields (HPF). Since our data did not follow normal distribution as assessed in Liljefors test, non-parametric method was applied. Results were subjected to Mann-Whitney U-test. p levels <0.05 were deemed statistically significant.
Results
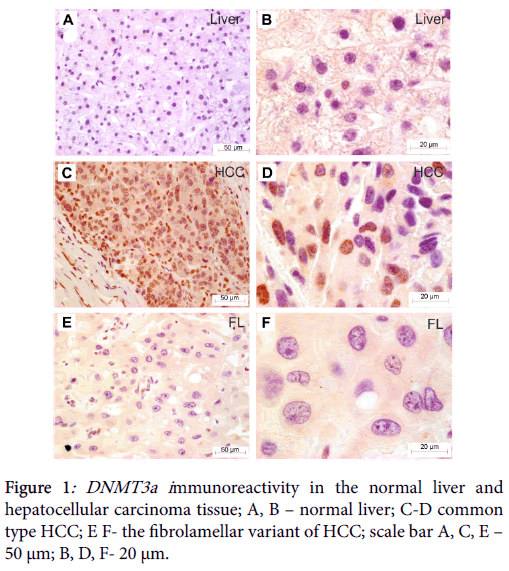
Immunohistochemical analysis revealed no immunostaining of DNMT3a in the non-neoplastic liver (Figure 1). In some hepatocytes weak cytoplasmic staining was observed. The analysis in cancer group has shown DNMT3a immunoreactivity to be predominantly localized in cancer cell nuclei in a form of separate large granules spotted in proximity to heterochromatin region (Figure 1).
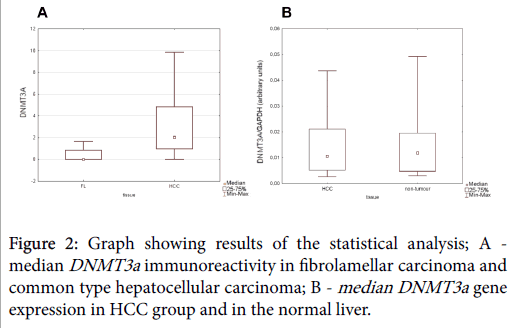
We found DMT3a immunoreactivity to be more abundantly expressed in non-fibrolamellar variant of HCC than in fibrolamellar hepatocellular carcinoma (U=10.5, p=0.0177; Mann-Whitney U test) (Figure 2).
No statistically significant difference in the intensity of immunostaining was detected between normal liver and HCC tissue sections. Similarly, gene expression analysis revealed no statistically significant difference between normal liver and HCC group (B; p>0.05 Mann-Whitney U test) (Figure 2).
Discussion
In our study we managed to confirm the previously proposed role of DNMT3a methyltransferase in pathogenesis of hepatocellular carcinoma. Although there was no quantitative difference between intensity of immunohistochemical reaction between non-neoplastic liver and HCC group as far as a whole of the cell is taken into account, the localization appears to be crucial for our results. DNMT3a can unfold its enzymatic activity with all its carcinogenic consequences in nucleus only, and that was indeed the localization of the protein in HCC tissues. On the other hand, the presence of DNMT3a in cytoplasm can have no influence on methylation status of the chromatin, therefore the cytoplasmic staining in non-neoplastic liver, although nearly identical in intensity to nucleic immunochemistry reaction in cancer, has indeed dramatically different biological significance.
This fact might be explained in terms of gene expression: although both HCC cells and normal hepatocytes produce DNMT3a mRNA in amounts that showed no significant difference in our study, the mRNA translation might be different or (and that is the explanation we find most probable) the cellular distribution, especially transport from cytoplasm into nucleus might be a crucial factor determining the biological activity on DNA of DNMT3a and explaining the differences in localisation.
However, the novel aspect of our study consisted chiefly in taking into account the fibrolamellar variant of HCC. In this case, it appeared that DNMT3a expression was significantly lower than in nonfibrolamellar variant of HCC. This finding is consistent with the observation that HBV, one of the most important risk factors globally for non-fibrolamellar type HCC, but not for FL induces DNMT3a expression [12]. Unfortunately, this difference could not be pin-point in terms of mRNA transcription due to lack of necessary fresh material with non-degraded RNA resulting from relative rarity of FL variant. Nonetheless this could be a promising direction for further research.
The relatively lesser role played by DNMT3a in progression of FL variant than in common type HCC elucidated in our study is a promising field for further analysis. First, it is clear than completely different molecular ethiopathogenesis of this specific variant extends to its epigenetic characteristics as well. Although, as noted in the introduction, DNMT3a is considered to be the main DNA methyltransferase involved in carcinogenesis of many tumours, it could not be ruled out that other enzymes of this family (DNMT3b and/or DNMT1) could have its say in FL variant. It has already been proved that DNMT1 and DNMT3a methylate preferentially different loci of the genome [15]. Thus it would not be surprising, if the lack of DNMT3a expression in terms of carcinogenesis could be replaced with DNMT1 overexpression leading to another outcome as far as diagnosis is concerned, i.e. progression to FL variant instead of common type HCC.
Our results suggest that an epigenetic DNA regulator, DNMT3a may have profound influence on hepatocarcinogenetic process. Taking into account the reduced presence of DNMT3a in the fibrolamellar variant of hepatocellular carcinoma it would be interesting to define if different epigenetic mechanisms are involved in this particular type of the liver cancer development. Improving our understanding of the roles of DNMT proteins in hepatocarcinogenesis can benefit in the development of epigenetic-based therapy designed for specific HCC subtype.
References
- Ozkul Y, Galderisi U (2016) The Impact of Epigenetics on Mesenchymal Stem Cell Biology. J Cell Physiol.
- Zoghbi HY, Beaudet AL (2016) Epigenetics and Human Disease. Cold Spring HarbPerspectBiol 8: a019497.
- Kisseljova NP, Kisseljov FL (2005) DNA demethylation and carcinogenesis. Biochemistry (Mosc) 70: 743-752.
- Feil R, Kelsey G (1997) Genomic imprinting: a chromatin connection. Am J Hum Genet 61: 1213-1219.
- Kohli RM, Zhang Y (2013) TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502: 472-479.
- Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, et al. (2015) The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J 34: 2350-2362.
- Kho MR, Baker DJ, Laayoun A, Smith SS (1998) Stalling of human DNA (cytosine-5) methyltransferase at single-strand conformers from a site of dynamic mutation. J MolBiol 275: 67-79.
- Xu F, Mao C, Ding Y, Rui C, Wu L, et al. (2010) Molecular and enzymatic profiles of mammalian DNA methyltransferases: structures and targets for drugs. Curr Med Chem 17: 4052-4071.
- Schoofs T, Müller-Tidow C (2011) DNA methylation as a pathogenic event and as a therapeutic target in AML. Cancer Treat Rev 37 Suppl 1: S13-18.
- Ding WJ, Fang JY, Chen XY, Peng YS (2008) The expression and clinical significance of DNA methyltransferase proteins in human gastric cancer. Dig Dis Sci 53: 2083-2089.
- Li XY, Feng XZ, Tang JZ, Dong K, Wang JF, et al. (2016) MicroRNA-200b inhibits the proliferation of hepatocellular carcinoma by targeting DNA methyltransferase 3a. Mol Med Rep 13: 3929-3935.
- Li H, Yang F, Gao B, Yu Z, Liu X, et al. (2015) Hepatitis B virus infection in hepatocellular carcinoma tissues upregulates expression of DNA methyltransferases. Int J ClinExp Med 8: 4175-4185.
- Waller LP, Deshpande V, Pyrsopoulos N (2015) Hepatocellular carcinoma: A comprehensive review. World J Hepatol 7: 2648-2663.
- Lim II, Farber BA, LaQuaglia MP (2014) Advances in fibrolamellar hepatocellular carcinoma: a review. Eur J PediatrSurg 24: 461-466.
- Fan H, Zhao Z, Cheng Y, Cui H, Qiao F, et al. (2015) Genome-wide profiling of DNA methylation reveals preferred sequences of DNMTs in hepatocellular carcinoma cells. Tumour Biol 37: 877-885.
Citation: Szparecki G, Ilczuk T, Wolosz D, Otto W, Gornicka B, et al. (2016) The Expression of DNA Methyltransferase DNMT3a in Classical and Fibrolamellar Hepatocellular Carcinoma. J Clin Exp Pathol 6:282. DOI: 10.4172/2161-0681.1000282
Copyright: © 2016 Szparecki G, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 12278
- [From(publication date): 6-2016 - Apr 02, 2025]
- Breakdown by view type
- HTML page views: 11301
- PDF downloads: 977