Research Article Open Access
The Establishment of Biosensor Technology Based on F0F1-ATPase Molecular Motor for Detection of Rotavirus and Hepatitis A Virus
Jie Zhang1*, Zhuo Zhao2, Mei-Ling Xu3, Xiang-Ying Yang1 and Zhi-Peng Liu2
1Beijing Inspection and Quarantine Testing Center, Beijing Entry-Exit Inspection Quarantine Bureau, Beijing, China
2Technical Center for Safety of Industrial Products, Tianjin Entry-Exit Inspection Quarantine Bureau, Tianjin, China
3Linyi Entry-Exit Inspection Quarantine Bureau, Linyi, China
- Corresponding Author:
- Zhang J, Beijing chaoyang
TianShuiYuan street no. 6, Beijing Inspection And Quarantine Testing Center
Beijing Entry-Exit Inspection Quarantine Bureau, Beijing, China, 100026
Tel/Fax: +86-10-58619231
E-mail: zhangjie@bjciq.gov.cn
Received Date: May 29, 2015 Accepted Date: August 18, 2015 Published Date: August 24, 2015
Citation: Zhang J, Zhao Z, Xu M, Yang X, Liu Z (2015) The Establishment of Biosensor Technology Based on F0F1-ATPase Molecular Motor for Detection of Rotavirus and Hepatitis A Virus. Biosens J 4:121. doi:10.4172/2090-4967.1000121
Copyright: © 2015 Zhang J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Biosensors Journal
Abstract
Around the world, food-borne virus is one of the main pathogenic microorganisms in the aspect of causing people and animals with acute diarrhoea and poses a serious threat to human health. Therefore, the rapid detection of foodborne virus is very important to guarantee food safety and human health. Here, we reported that we developed a specific, convenient and fast detected method to detect rotavirus (RV) and hepatitis A virus (HAV) by using F0F1-ATPase molecular motor biosensor. Specific RNA probes were encompassed the conservative region of food-borne virus, and a molecular motor detect device was constructed by connecting probes to F0F1-ATPase molecular motor through biotin-streptavidin system. Extracted virus RNA was conjugated with the biosensor separately and meanwhile ATP was synthesized. By comparing their fluorescence intensity, virus RNA level was detected. Our results demonstrated that this biosensor’s sensitivity was the concentrations of 0.005 ng/mL and 0.01 ng/mL for RV and HAV respectively. Furthermore, this method possessed specificity for RV and HAV and none cross-reaction between them. What’s more, this method could be accomplished within 1h. We detected 15 samples by using this method and the results were consistent with RT-PCR results. Overall, this new-typed method based on F0F1-ATPase molecular motor biosensor for RV and HAV detection is sensitive and specific and can be used in the rapid detection of food-borne virus.
Keywords
Food-borne virus; Molecular motor biosensor; Detection; F0F1-ATPase
Introduction
Food-borne virus, such as human calicivirus, rotavirus, astrovirus, the enteric adenovirus, hepatitis A virus, norovirus, is mainly transmitted through the route of contaminated food, water, and life, and poses a serious threat to human health [1-7]. For the reason of food safety, the rapid detection for food-borne virus is very important for food safety and human health. The traditional method of foodborne virus detection mainly includes electron technology, cultivation, immune detection and gene detection technology [8-11]. But, with the requirements of rapid detection of clinical research and importexport trade, the conventional detection methods have been unable to meet. Therefore, it is necessary to develop a more sensitive and specific detection methods for rapid detection of food-borne virus. Molecular motor biological sensing technology is a new kind of technology developed in the recent years and it has intuitive, high sensitivity, high speed, easy operation and less pollution properties. But up to now, the reports of molecular motor biosensor have been still limited in the application of food-borne virus detection.
F0F1-ATPase, one of important molecular motor, is a rotating biological molecular motor and is responsible for the biological energy conversion in vivo [12,13]. F0F1-ATPase consists of two parts, F0(ab2cn) and F1(α3β3γδε), and they are embedded in the membrane and prominent in membrane respectively [14-16]. ATP synthase can synthesize ATP through using trans membrane proton and can also transport protons through the hydrolysis of ATP. In the process of synthase of ATP, protons can be pumped from inside membrane to outside membrane of Chromatophore. This progress could make the change of H+ concentration in the endometrial microenvironment solution [17,18]. If F0F1-ATPase molecular motor is design into a nano device and this nano device can transform the chemical energy into mechanical energy, then F0F1-ATPase molecular motor biosensor will be a newly detected platform for detection of pathogenic microorganisms. In present, the existing F0F1-ATPase molecular motor biosensor is mainly constructed by conjugating specific molecular probes through biotin-avidin system and has been used to detect many of pathogens and viruses such as bird flu virus, mouse hepatitis virus, salmonella spp., vibrio cholera- [19-22]. But, the applications of F0F1- ATPase molecular motor biosensor in the field of virus detection are still limited.
Presently, in order to improve the technological level to detection and strengthen the prevention and control of food-borne virus, we have developed a novel F0F1-ATPase molecular motor biosensor detection method for RV and HAV detection and it exhibits excellent performance [23,24]. We propose this research might provide better solutions in the application of rapid detection of food borne virus.
Materials and Methods
Chemicals, materials and instruments
All chemicals that used in the experiments were reagent grade and were used as received following; glycerol, Magnesium chloride hexahydrate (MgCl2•6H2O), Tricine, Potassium phosphate monobasic (KH2PO4), Sodium chloride (NaCl), Potassium chloride (KCl) and Potassium phosphate dibasic (K2HPO4) were purchased from Sigma- Aldrich Chemical (St. Louis, MO, USA).
Freeze-dried live attenuated hepatitis a vaccine was purchased from Zhejiang Pukang biotechnology co., LTD; rotavirus was provided by National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention; Biotin-AC5-Sulfo-Osu was purchased from Dojindo Laboratories (Kumamoto, Japan). Streptavidin and adenosine diphosphate (ADP) were purchased from Sigma (St. Louis, MO, USA). N-(fluorescein-5-thiocarbamoyl)- 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine, triethylammonium salt (F-DHPE) was purchased from invitrogen (Carlsbad, CA, USA).
All fluorescence test experiments were performed using SynergyTM 4 multi-detection microplate reader (BioTek UK, Potton, UK).
RNA extraction
The total mRNA of RV and HAV was extracted by using the RNA extraction kit (Qiangen, Valencia, CA, USA) according to the manufacturer's instructions and RNA sample was used as the target of the molecular motor biosensor.
Synthesizing and labeling of probes
Based on bioinformatics analysis, we designed the probes respectively for RV and HAV, and sequences are following as: RV, 5’- AAGCGGATTATGCAGAAGCACTG -3’; HAV, 5’-AGCGGCGGATATTGGTGAGTTGTTAAGAC-3’. The probe’s location of RV is in the highly conserved region of VP7 gene. The sequence of HAV’s probe is a consensus region and highly conserved in all gene phenotypes of hepatitis A virus. All probes were labeled by biotin in the 5’ end and purchased from Takara (Kyoto, Japan).
Preparation of chromatophore and labeling F-DHPE
Chromatophores were prepared from the cells of Thermomicrobium roseum (T. roseum) according to the Refs. [12,25]. F-DHPE, a fluorescence probe, could be labeled onto the surface of Chromatophores [12]. In our study, 10 μL aliquots of F-DHPE (200 mg/mL, dissolved in ethanol) were mixed with 200 μL of Chromatophores, and incubated for 15 min in dark with gentle shaking at room temperature. After incubation, labeled Chromatophores were harvested by centrifugation at 30, 000 r/min (4°C for 15 min) and free F-DHPE was washed away with 10 mM PBS by centrifugation at 10, 000 r/min at 4°C for 15 min three times. The resulting pellets were re-suspended with 200 μL of PBS. The labeled Chromatophores are called fluorescent Chromatophores in the following text.
Construction of F0F1-ATPase molecular motor biosensor
Firstly, 2 μL of biotin (biotin-AC5-Sulfo-Osu, 2 μM) was added into 20 μL of ε-subunit antibody and the mixture was incubated 30 min at room temperature. In this step, the N terminal of antibody was marked. Next, the F0F1-ATPase molecular motor biosensor was constructed as follows: 40 μg aliquots of biotin-labeled ε-subunit antibody were added into 200 μL of fluorescent Chromatophores and PBS was added to a final volume of 1 mL. After incubating at 37°C for 1 h, PBS was added to a final volume of 1.4 mL. The pellets were harvested by centrifugation at 30,000 g at 4°C for 10 min and resuspended with 500 μL of PBS. 2 μL of avidin (2 mg/mL) and PBS were added into a final volume of 1 mL. The mixture was shaken at 50~100 rpm at room temperature for 10 min. PBS was added to a final volume of 1.4 mL. The pellets were harvested by centrifugation at 30,000 g at 4°C for 10 min and resuspended with 500 μL of PBS. Subsequently, a volume of 2 μL (2 μM) of biotin-labeled probe and PBS were added to a final volume of 1 mL. The mixture was shaken at 50~100 rpm at room temperature for 10 min, PBS was added to a final volume of 1.4 mL., The pellets were harvested by centrifugation at 30,000g at 4°C for 10 min and Chromatophores were resuspended with 150 μL of glycerol (30%, V/V). The prepared F0F1-ATPase molecular motor biosensors were called ChroRV and ChroHAV, and stored at −20°C following test.
Construction of molecular motor detection method
ChroRV and ChroHAV were diluted to a certain ratio by using synthetic buffer (glycerol 20%, MgCl2 5 mM, Tricine 50 mM, KH2PO4 5 mM). 10 μL of samples was added to this mixes and blended through vortex shocking. Water and/or synthetic buffer were used as the blank. After adding 30 μL of start buffer (start buffer was prepared with 1.6 M ADP and synthetic buffer (1:3, v/v) before use), the reaction system was incubated 10 min at room temperature. Add 200 μL of PBS, and detect the fluorescence signal by using 96 well plates by microplate reader. The results were calculated from fluorescence value by the formula of
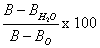
(B: sample fluorescence, B0: background values, BH2O: fluorescence of water).
Statistical analyses
All experimental data were shown as the mean ± standard deviation. Statistical significance was evaluated with SPSS software. Experimental differences of multiple groups (>3 groups) were analyzed using the two-tailed Student’s t-text. P values<0.05 was considered statistic significant.
Results and Discussion
The construction of F0F1-ATPase molecular motor biosensor
F-DHPE is known as a lipid fluorescent probe and can be embedded into the phospholipid molecules layer [12]. Furthermore, F-DHPE is a pH indicator and has been used to measure pH changes of phospholipid layer outside through embedding into lipid bilayer. In the range of pH 7.0 to 9.0, the fluorescence intensity of F-DHPE has a positive correlationship with PH [12]. In our study, F-DHPE was labeled into the phospholipid molecules layer to point out the synthetic efficiency of ATP. F0F1-ATPase is a rotary motor. During ATP synthesis, protons are pumped out of the Chromatophores, resulting in an increase of concentration of H+ out of the Chromatophores. Thus, the pH out of the Chromatophores will decrease, and the fluorescence probe F-DHPE is expected to detect the pH decrease [12]. In present, we successfully connected “ε-subunit antibody-streptavidin-biotin-probe” system to F0F1-ATPase and constructed the F0F1-ATPase molecular motor biosensor (Supplementary Figure S1). Generally, fluorescence intensity is higher significantly than that of without loads. However, because of Brownian motion, the fluorescence intensity is often lower than that of without loads. In other words, the significant differences in fluorescence intensity compared with negative control suggest the successful capture of target molecule.
F0F1-ATPase, a mitochondrial F0F1-ATP synthase, has a unique property due to its quickly rotary characteristics [12,13]. As we know, the F0F1 motor has many rotary subunits such as α-subunits, β-subunits and ε-subunits. Some reports showed that the ε-subunits may be more sensitive to the binding load than the other subunits [26,27]. According to these properties of F0F1 motor, in our study, we establish the coupling between the catalytic site (ε-subunits) and proton transferring. Additionally, conventional molecular experiments have many of operations such as PCR AMPLICATION, electrophoresis and hybridization; Due to large reduction of these usual operations the molecular motor biosensor greatly reduced the likelihood of contamination [28]. Molecular motor biosensor has rapid and precise properties and if we can make it subminiature and portable, or make it directly to detect at the scene of sample, it could be wildly applied. Predictably, along with the progress of micromachining technology and nanotechnology [29-32], the molecular motor biosensor will be largely applied in the various detection fields.
Establishment of the molecular motor biosensor reaction system
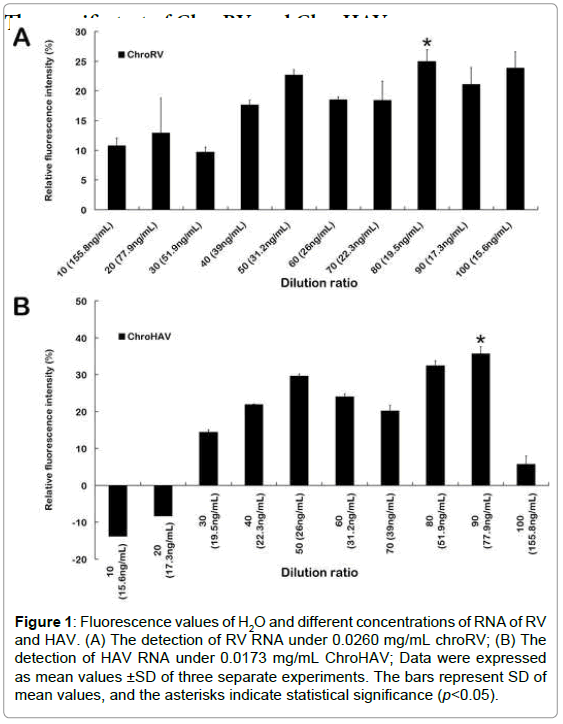
Firstly, ChroRV and ChroHAV were diluted by concentration gradient. In the different concentration conditions, the fluorescence value of H2O and virus was detected, and the fluorescence differentials were shown as Figure 1. From the results, we found that the dilution radio is lower and the fluorescence intensity of final reaction system is higher. Because along with the concentration of molecular motor biosensor increasing, the reaction system has more ATPase, and therefore fluorescence intensity is larger. Based on the principle of maximum differentials between molecular motor and H2O, we finally chose the 0.015 mg/mL, 0.0173 mg/mL and 0.0260 mg/mL as the final concentration of ChroRV and ChroHAV respectively. Meanwhile, we have explored the optimum reaction concentrations of all three virus RNA and data was shown as Supplemental Figure S2. We found that the fluorescence values of different concentration of RNA were all higher than the control (H2O), especially 0.6 ng/ μL and 0.9 ng/μL of RV and HAV respectively indicating that the detection of F0F1-ATPase molecular motor biosensor is effective and the optimum concentration of RNA is 0.6 ng/μL and 0.9 ng/μL of RV and HAV respectively.
Figure 1:Fluorescence values of H2O and different concentrations of RNA of RV and HAV. (A) The detection of RV RNA under 0.0260 mg/mL chroRV; (B) The detection of HAV RNA under 0.0173 mg/mL ChroHAV; Data were expressed as mean values ±SD of three separate experiments. The bars represent SD of mean values, and the asterisks indicate statistical significance (p<0.05).
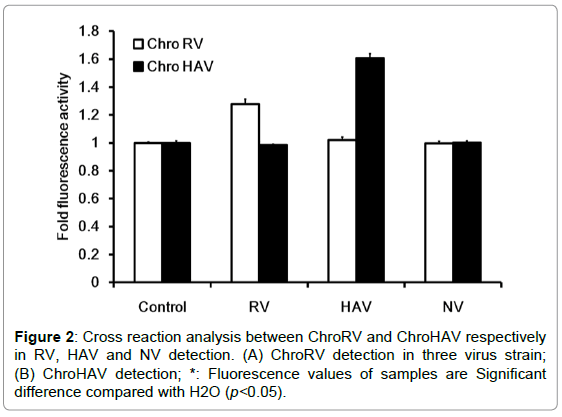
In order to detect whether there were cross reactions of F0F1- ATPase molecular motor biosensor, we furthermore tested the specificity of ChroRV and ChroHAV respectively in RV, HAV and norovirus (NV) detection and data was shown as Figure 2. In RV detection, the molecular-biosensor fluorescence intensity of RV was significantly higher than the fluorescence intensity of H2O, however, the fluorescence intensity of HAV and NV was basically unchanged compared with the control indicating that the molecular motor biosensor of ChroRV is specific RNA and has none cross reactions with HAV and NV. Similarly, we have drawn the same conclusion in the specific test of HAV detection. Together, above result showed that F0F1-ATPase molecular motor biosensor (ChroRV and ChroHAV) is specific to the RV and HAV detection and there is no cross detection.
The sensitive test of F0F1-ATPase molecular motor
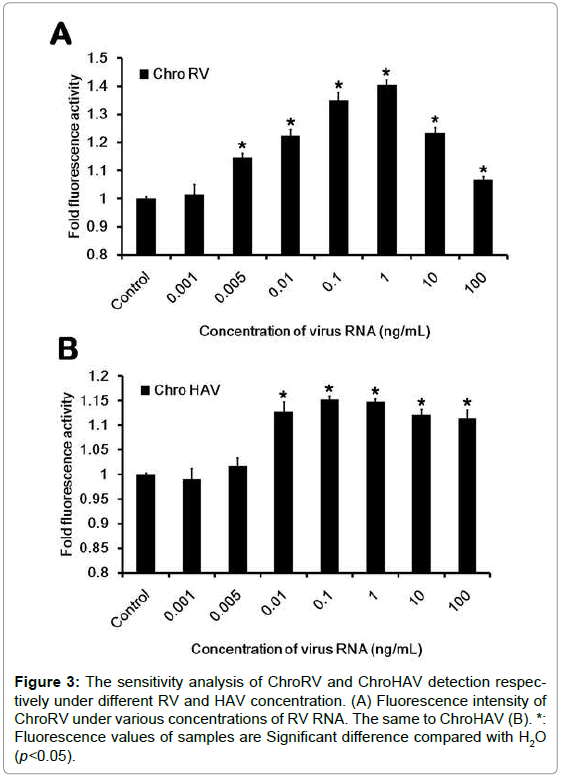
To furthermore detect the sensitivity, we have next designed and performed the sensitive test. We first carried on the concentration gradient dilution of virus RNA and the concentration was respectively diluted to the 0.001 ng/mL, 0.005 ng/mL, 0.01 ng/mL, 0.1 ng/mL, 1 ng/ mL, 10 ng/mL and 100 ng/mL. After dilution, the sensitive test was carried out and data was shown as Figure 3. In RV detection, we found that when the concentration is in the range of 0.005~100 ng/mL, the fluorescence of ChroRV is significantly higher than the control, but the concentration of 0.001 ng/mL indicating that the sensitivity of ChroRV is the concentration of 0.005 ng/mL. Similarly, we next demonstrated the detected sensitivity of ChroHAV is the concentration of 0.01 ng/mL.
Figure 3:The sensitivity analysis of ChroRV and ChroHAV detection respectively under different RV and HAV concentration. (A) Fluorescence intensity of ChroRV under various concentrations of RV RNA. The same to ChroHAV (B). *: Fluorescence values of samples are Significant difference compared with H2O (p<0.05).
As we know, the concentration of food-borne virus in food is usually lower and meanwhile the sample amount of tested food has a large quantity. Additionally, the pathogenic quantity of food-borne virus infection is lower and especially 10~100 units of virus can cause infection. Therefore, a new-typed, high effective, rapid, precise and sensitive detection method is very necessary to the supervision of import-export detections. In this background, the establishment of our new detected method has been performed. Here, we just established the molecular motor detection in RV and HAV, but the usual food-borne virus has many kinds, such as human calicivirus (HuCV), astrovirus (AV) and enteric adenovirus (EV). Therefore, based on this study we next expanded the scope of food-borne virus detection and tried to simultaneously detect various food-borne viruses.
Applications of F0F1-ATPase molecular motor in the sample testing
In the above experiments, we proved that F0F1-ATPase molecular motors for the detection of RV and HAV were effective and specific, but the effectivity of biosensor in the applications of actual samples testes is still unknown. To address this question, we randomly chose 15 known negative/positive samples as the tested samples to perform molecular motor biosensor detection and the results were compared with the results of RT-PCR detected method (Table 1 and Supplemental Figure S3). From the result, we found that the result of molecular motor detection was absolutely same as the result of RT-PCR detection in RV and HAV detection respectively indicating that the detection of ChroRV and ChroHAV is completely used for the food-borne viruses daily inspection. The fluorescence value of molecular motor biosensor and control was shown as Supplemental Figure S3. Food-borne virus infection is increasingly becoming a public health problem in the worldwide [1]. Therefore, the establishment of sensitive and rapid detected method for food-borne virus detection is of great significance for protecting human health and preventing the outbreaks. Our result demonstrated the practical application of F0F1-ATPase biosensor and we hope these findings could be helpful to virus prevention and control.
| Virus | Methods | Samples No. | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | ||
| RV | RT-PCR | ï¼�? | ï¼�? | ï¼�? | ï¼ | ï¼ | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? |
| Molecular motor | ï¼�? | ï¼�? | ï¼�? | ï¼ | ï¼ | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | |
| HAV | RT-PCR | ï¼�? | ï¼ | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼ | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? |
| Molecular motor | ï¼�? | ï¼ | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼ | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | ï¼�? | |
Table 1: The comparative results between RT-PCR detection and molecular motor detection.
Conclusion
In present, we have successfully used the Chromatophores extracted from thermophilic bacteria to construct a newly typed molecular motor biosensor and made it apply to RV and HAV detection. In our studies, we systematically investigated the specificity, sensitivity and veracity of F0F1-ATPase biosensor and the results revealed that it is an effective detected method for the detection of food-borne virus. What’s more, the significance of our study is that a rapid and sensitive detected method is provided for the food-borne virus detection and the test time can be shortened within 1 h. Additionally, due to large preparation of F0F1-ATPase molecular motors biosensor, the cost of this method is lower than that of other detection. We propose these findings might provide better solutions for clinical diagnosis, food detection and disease prevention of food-borne virus in the future.
Acknowledgement
We are grateful to Prof. Jiachang Yue, Institute of Biophysics, Chinese Academy of Sciences for his valuable suggestions.
Author Contributions
Jie Zhang and Zhuo Zhao designed the study and interpreted the results. Mei- Ling Xu, Xiang-Ying Yang and Zhi-Peng Liu collected test data and drafted the manuscript.
References
- Appleton H (2000) Control of food-borne viruses. Br Med Bull 56: 172-183.
- Bouzalas IG, Wuthrich D, Walland J, Drogemuller C, Zurbriggen A, et al. (2014) Neurotropic astrovirus in cattle with nonsuppurative encephalitis in Europe. J ClinMicrobiol52: 3318-3324.
- Gomez-Santiago F, Ribas-Aparicio RM, Garcia-Lozano H (2012) Molecular characterization of human calicivirus associated with acute diarrheal disease in Mexican children. Virol J 9: 54.
- Grondahl-Rosado RC, Yarovitsyna E, Trettenes E, Myrmel M, Robertson LJ (2014) A One Year Study on the Concentrations of Norovirus and Enteric Adenoviruses in Wastewater and A Surface Drinking Water Source in Norway. Food Environ Virol.
- Parashar UD, Li JF, Cama R, DeZalia M, Monroe SS, et al. (2004) Human caliciviruses as a cause of severe gastroenteritis in Peruvian children. J Infect Dis 190: 1088-1092.
- Stefkovicova M, Litvova S, Simurka P, Goczeova J, Gajdosikova A, et al. (2015) Rotavirus type profile in nosocomial and community infections in Western Slovakia. Folia Microbiol (Praha) 60: 177-181.
- Wang X, Ren J, Gao Q, Hu Z, Sun Y, et al. (2015) Hepatitis A virus and the origins of picornaviruses. Nature 517: 85-88.
- Dea S, Garzon S (1991) Identification of coronaviruses by the use of indirect protein A-gold immunoelectron microscopy. J Vet Diagn Invest 3: 297-305.
- Lozano LF, Hammami S, Castro AE, Osburn B (1992) Comparison of electron microscopy and polyacrylamide gel electrophoresis in the diagnosis of avian reovirus and rotavirus infections. Avian Dis 36: 183-188.
- Saif LJ, Bohl EH, Kohler EM, Hughes JH (1977) Immune electron microscopy of transmissible gastroenteritis virus and rotavirus (reovirus-like agent) of swine. Am J Vet Res 38: 13-20.
- Sugihara K, Reupke H, Schmidt-Westhausen A, Pohle HD, Gelderblom HR, et al. (1990) Negative staining EM for the detection of Epstein-Barr virus in oral hairy leukoplakia. J Oral Pathol Med 19: 367-370.
- Cui Y, Zhang F, Yue J (2005) Detecting proton flux across chromatophores driven by F0F1-ATPase using N-(fluorescein-5-thiocarbamoyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolami ne, triethylammonium salt. Anal Biochem344: 102-107.
- Shu YG, Yue JC, Ou-Yang ZC (2010) F0F1-ATPase, rotary motor and biosensor. Nanoscale 2: 1284-1293.
- Fillingame RH (1997) Coupling H+ transport and ATP synthesis in F1F0-ATP synthases: glimpses of interacting parts in a dynamic molecular machine. J ExpBiol200: 217-224.
- Stock D, Leslie AG, Walker JE (1999) Molecular architecture of the rotary motor in ATP synthase. Science 286: 1700-1705.
- Yasuda R, Noji H, Yoshida M, Kinosita KJ, Itoh H (2001) Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410: 898-904.
- Gao YQ, Yang W, Karplus M (2005) A structure-based model for the synthesis and hydrolysis of ATP by F1-ATPase. Cell 123: 195-205.
- Sabbert D, Engelbrecht S, Junge W (1996) Intersubunit rotation in active F-ATPase. Nature 381: 623-625.
- Elston T, Wang H, Oster G (1998) Energy transduction in ATP synthase. Nature 391: 510-513.
- Kinosita KJ, Yasuda R, Noji H, Adachi K (2000) A rotary molecular motor that can work at near 100% efficiency. Philos Trans R SocLond B BiolSci355: 473-489.
- Noji H, Yasuda R, Yoshida M, Kinosita KJ (1997) Direct observation of the rotation of F1-ATPase. Nature 386: 299-302.
- Yasuda R, Noji H, Kinosita KJ, Yoshida M (1998) F1-ATPase is a highly efficient molecular motor that rotates with discrete 120 degree steps. Cell 93: 1117-1124.
- Zhang J, Wang X, Xu M, Zhang H, Zhang L, et al. (2013) F0F1-ATPase Molecular Motor Biosensor for Rapid Detection of Hepatitis A Virus in Foods. Food Science: 178-182.
- Zhang J, Xu M, Wang X, Wang Y, Wang X, et al. (2013) [Detection of food-borne rotavirus by molecular motor biosensor]. Sheng Wu Gong Cheng XueBao 29: 681-690.
- Suzuki T, Murakami T, Iino R, Suzuki J, Ono S, et al. (2003) F0F1-ATPase/synthase is geared to the synthesis mode by conformational rearrangement of epsilon subunit in response to proton motive force and ADP/ATP balance. J BiolChem278: 46840-46846.
- Cheng J, Zhang XA, Shu YG, Yue JC (2010) F0F1-ATPase activity regulated by external links on beta subunits. BiochemBiophys Res Commun 391: 182-186.
- Zhao Y, Wang P, Wang F, Zhou H, Li W, et al. (2012) A novel biosensor regulated by the rotator of F0F1-ATPase to detect deoxynivalenol rapidly. BiochemBiophys Res Commun 423: 195-199.
- Zhang J, Li Z, Zhang H, Wang J, Liu Y, et al. (2013) Rapid detection of several foodborne pathogens by F0F1-ATPase molecular motor biosensor. J Microbiol Methods 93: 37-41.
- Atar N, Eren T, Yola ML, Wang S (2015) A sensitive molecular imprinted surface plasmon resonance nanosensor for selective determination of trace triclosan in wastewater. Sensors and Actuators B: Chemical 216: 638-644.
- Maleh HK, Javazmi FT, Atar N, Yola ML, Gupta VK, et al. (2015) A Novel DNA Biosensor Based on a Pencil Graphite Electrode Modified with Polypyrrole/Functionalized Multiwalled Carbon Nanotubes for Determination of 6-Mercaptopurine Anticancer Drug. IndEngChem Res 54: 3634-3639.
- Atar N, Eren T, Yola ML (2015) A molecular imprinted SPR biosensor for sensitive determination of citrinin in red yeast rice. Food Chem184: 7-11.
- Atar N, Eren T, Yola ML, Maleh HK, Demirdogen B (2015) Magnetic iron oxide and iron oxide@gold nanoparticle anchored nitrogen and sulfur-functionalized reduced graphene oxide electrocatalystfor methanol oxidation. RSC Advances 5: 26402-26409.
Relevant Topics
- Amperometric Biosensors
- Biomedical Sensor
- Bioreceptors
- Biosensors Application
- Biosensors Companies and Market Analysis
- Biotransducer
- Chemical Sensors
- Colorimetric Biosensors
- DNA Biosensors
- Electrochemical Biosensors
- Glucose Biosensors
- Graphene Biosensors
- Imaging Sensors
- Microbial Biosensors
- Nucleic Acid Interactions
- Optical Biosensor
- Piezo Electric Sensor
- Potentiometric Biosensors
- Surface Attachment of the Biological Elements
- Surface Plasmon Resonance
- Transducers
Recommended Journals
Article Tools
Article Usage
- Total views: 12594
- [From(publication date):
specialissue-2015 - Apr 02, 2025] - Breakdown by view type
- HTML page views : 11631
- PDF downloads : 963