Research Article Open Access
The Effects of DNA Methylation on the Expression of Non-imprinted Genes in Rice
Hongyu Zhang*, Yutong Liu, Mipeng Han, Limei Wu, Zhijian Liu, Xiaotong Chen, Peizhou Xu and Xianjun WuRice Research Institute, Sichuan Agricultural University, Wenjiang, Chengdu, P.R.China
- Corresponding Author:
- Hongyu Zhang
Gulf Coast Research and Education Center
University of Florida, USA
Tel: +8613708203775
Fax: +862886290897
E-mail: wuxjsau@126.com
Received date: March 08, 2016; Accepted date: April 13, 2016; Published date: April 18, 2016
Citation: Zhang H, Liu Y, Han M, Wu L, Liu Z, et al. (2016) The Effects of DNA Methylation on the Expression of Non-imprinted Genes in Rice. J Rice Res 4:168. doi:10.4172/2375-4338.1000168
Copyright: © 2016 Zhang H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Rice Research: Open Access
Abstract
The expression of imprinted genes is regulated by well-known genetic mechanisms such as DNA and histone methylation. However the mechanism regulating the expression of non-imprinted genes that are specifically expressed in endosperm is currently unknown. To determine whether DNA methylation is involved in the regulation of non-imprinted gene expression in endosperm, we used rice seeds from a reciprocal cross between cultivars Nipponbare and 9311 treated with a methylation inhibitor 5-aza-2’-deoxycytidine to investigate the expression patterns of four non-imprinted genes in seedlings. We found these endosperm specific genes were activated in F1 with two types of expression patterns: 1) either both parental alleles were expressed in F1 leaves; 2) or only one parental allele was expressed in the leaves of the progeny. We speculate that the altered expression patterns of parental alleles may be associated with F1 heterosis. We also observed that the expression of non-imprinted genes underwent dynamic changes at different development stages with two showing imprinted expression patterns, suggesting that DNA methylation is involved in regulating the expression of some imprinted as well as non-imprinted genes. The results of this study provide a reference for further exploring epigenetic mechanisms underlying seed development and, potentially, the association of dynamic changes of DNA methylation with heterosis.
Keywords
Rice; Imprinted genes; Non-imprinted genes; Endosperm; 5-azadC; DNA methylation
Introduction
Cereal grain consists of two major components: embryo and endosperm that are the products of double fertilization specifically occurring in flowering plants. Endosperm occupies most of the volume of the grain and is a major source of human nutrition. Thus it is important to investigate grain development to improve crop and food production to fulfill the growth of world population.
Genomic imprinting almost exclusively occurs in plant endosperm and is modulated by the variation of DNA and histone methylation [1-7]. Gene imprinting refers to the phenomenon that one of the parental gene copies is exclusively or preferentially expressed depending on parent-of-origin [8-11]. Proper endosperm development requires a set of genes being imprinted and these genes may affect grain size [12]. The imbalance of parental genome dosage in the endosperm observed in interploidy crosses and DNA methylation mutants is the main reason for endosperm abortion [13-18]. Endosperm development is regulated by epigenetic mechanisms such as DNA and histone methylation that regulate the expression of imprinted genes [19-27]. Because endosperm is a terminally differentiated tissue, the genetic and epigenetic variations that occur in imprinted genes will not be passed to the generation.
Coffspringhanges of DNA methylation patterns and levels are closely related to the formation of heterosis [28-33]. The patterns of plant DNA methylation are tissue- and organ-specific [34,35]. For example, elevated DNA methylation levels at some loci are considered to be positively associated with plant heterosis [32,36], while at other loci it is the decreased DNA methylation levels that correlates with the plant heterosis [31,37]. Specifically expressed genes in endosperm may influence embryonic development via influencing endosperm development [22]. Ectopic activation of these endosperm specific genes in seedlings or other organs may impact on plant growth and thereby heterosis. Zemach [38] identified 165 endosperm-specific genes using gene chip technology and found that not all were imprinted genes. Luo et al. [23] identified 262 candidate imprinted loci in rice by Illumina high-throughput sequencing and verified 56 imprinted loci in the endosperm. These authors identified a few imprinted rice candidates that are highly homologous to genes in Arabidopsis thaliana associating with epigenetic regulation, including DNA methylation, histone methylation, and small RNA pathways. Wang et al. [39] found that the expression of genes silenced by DNA methylation could be activated through the addition of the methylation inhibitor 5-aza-2’- deoxycytidine (5-azadC) [40]. However, it is still unknown whether DNA methylation is involved in the regulation of non-imprinted genes during endosperm development.
In this study, we treated the seeds of a hybrid rice (Oryza sativa) from a reciprocal cross between japonica cultivar Nipponbare and indica cultivar 9311 with the DNA methylation inhibitor 5-azadC (thereby altering the expression of genes in the resulting seedlings). We then analyzed the regulatory effect of DNA methylation on nonimprinted genes. The results of the present study provide a reference for further elucidating the genetic mechanism underlying heterosis and the molecular mechanism of imprinting during endosperm development.
Materials and Methods
Plant materials
Rice (Oryza sativa) line Parents Nip (Nipponbare) line 9311, planted in Sichuan Wenjiang test fields with reciprocal cross in the flowering phase, taking the seed endosperm with the development of 3-5D and the parental roots, stems, endosperm, leaves, freezing in liquid nitrogen and preserving at -80°C to prepare RNA extraction.
The mature hybrid seeds were collected and preserved at -4°C for one month, to break seed dormancy for the preparation of the subsequent pharmaceutical treatment and germination. RNA isolation, reverse transcription and sequencing.
Total RNA was isolated using the RNeasy Plant Mini kit (Qiagen, Hilden, Germany) according manufacturers’ instruction. The genomic DNA contamination was removed by using RQ1 RNase-Free DNase (Promega, Madison, WI). cDNA was synthesized from the 100 ng of total RNA samples by using the PrimeScript First Strand cDNA Synthesis kit (Takara, Dalian, Chian). After synthesizing cDNA, PCR amplification was performed using the TaKaRa PCR Amplification kit (Takara) using the primers listed in Table 1. The PCR condition was 95°C for 3 min for pre-denaturing, and then 34 cycles for denaturing at 72°C for 45S, annealing at 56°C and polymerization at 72°C for 7 min. PCR products were examined on a 1% agarose gel followed by sequencing in BGI (Shenzhen, China).
| Gene Name | Gene Annotation | Primer sequences(5’-3’) |
|---|---|---|
| LOC_Os01g01290 | histone-like transcription factor and archaeal histone, putative, expressed (CCAAT Family) | CACCAAAGGCTCAACAACAA |
| ACGGTTATGGGATTGAGCAG | ||
| LOC_Os01g01470 | no apical meristem protein, putative, expressed (NAC Family) | TGGGTCATGCACGAGTACAG |
| AGTGAGAGTGAAGCGGTGGT | ||
| LOC_Os01g24460 | histone-like transcription factor and archaeal histone, putative, expressed (CCAATFamily) | GGCCAAGAAGAACAACATGAG |
| GCCATTACTGGTGCTTGGAT | ||
| LOC_Os01g29840 | no apical meristem protein, putative, expressed (NAC Family) | CAGATGCCCTCCATGTCTG |
| GACACCACCAGCGACGAC | ||
| LOC_Os01g33350 | Expressed protein | GAATTTGTGCCTCCATGGTT |
| CGTGGTATGATGATCGCCTA | ||
| LOC_Os01g39850 | histone-like transcription factor and archaeal histone, putative, expressed (CCAAT Family) | CCATTGCCTCCACAGAGTC |
| GACCCCTTGCTATGTTGTGAA | ||
| LOC_Os02g12310 | no apical meristem protein, putative, expressed (NAC Family) | TCTTCGGTGAATCGTCTTCC |
| CCACCATGGTTTCTTTGCAT | ||
| LOC_Os02g15350 | Transcriptional activator can collaborate LOC_Os07g08420 regulate seed storage protein (SSP)gene expression (C2C2-DOF Family) in Rice | ATTATCCCCGGTGGAGGAG |
| CAGGAGCAGGAGGAGACG | ||
| LOC_Os04g10260 | bZIP transcription factor basic region leucine zipper domain containing protein | TCAGTTAAGCCGGAGGTCAC |
| TGAATTTCACATTCGCAAGC | ||
| LOC_Os04g35010 | helix-loop-helix DNA-binding domain containing protein, expressed (BHLH Family) | TCTGGTAAGGTCGATTAAAGCA |
| CATCTTCTTCCTCCGCTCTC | ||
| LOC_Os05g34310 | no apical meristem protein, putative, expressed (NAC Family) | CTTCAACCCGTGGGAGCTT |
| CGAGCACTGTAACCGTGAGA | ||
| LOC_Os07g08420 | bZIP transcription factor,Transcriptional activator can collaborate LOC_Os02g15350regulate seed storage protein (SSP)gene expression (C2C2-DOF Family) in Rice | ACAACTTCACCAGGCCATTC |
| GCTCCATGTTGACAAGCTCA | ||
| LOC_Os09g34880 | basic region leucine zipper domain containing protein, expressed (bZIP Transcription Factor) | CTCCCTTCCTCGGTCCTCT |
| TTTGGCTGTGGAAACCCTAC | ||
| LOC_Os10g25850 | Nuclear transcription factor Y subunit A-7 | AAACCTGAGTGCAACCAACC |
| ATGCCTCAATTTTGCTTGCT | ||
| LOC_Os11g31330 | no apical meristem protein | TCGGAGGTGCCCATCTATTA |
| AGGGTGGCTCTGAACCATT | ||
| LOC_Os11g31340 | no apical meristem protein, putative, expressed (NAC Family) | CTGCTGGTGATGGGTTCTG |
| GCGATGGTCGTTCCTGTG | ||
| LOC_Os11g31360 | no apical meristem protein, putative, expressed conserved seed development associated transcription factors, arabidopsis its analogues function is to regulate embryo (NAC Family) | CAAGGAGAACAGCCACCCTA |
| CATGAGTATGGGCAGCAGAC | ||
| LOC_Os11g31380 | no apical meristem protein | CTCAAAACCACCCTGCAACT |
| CTCCCTTGCATTGCCATT |
Table 1: Preliminary screening, gene annotation, and primer sequences.
Gene annotation and primer sequences
According to the gene annotation provided by http://bioinformatics.cau.edu.cn/ neweasygo/ to confirm the genes encoding genes encoding transcription factors in Assaf According to http://blast.ncbi.nlm.nih.gov/ to detect the differences of polymorphic loci between japonica rice and indica rice, using Primer Premier 5 to design the primers. A total of 18 genes were identified.
Seed treatment by methylation inhibitor 5-azadC
The concentration of 5-azadC was diluted to 30, 50 μmol/L dissolving with DMSO, diluting with distilled water to the corresponding concentration; the control group was a mixed solution of DMSO and distilled water. The seeds were steeped in the condition of light incubator temperature (28°C/25°C) and photoperiod(16h D/8h N) for 3-5d,observing the changes of plant properties after treatment according to the germination, The drug concentrations used finally was confirmed to be 3 μmol/L.
Results
Identification of the genes specifically expressed in endosperm
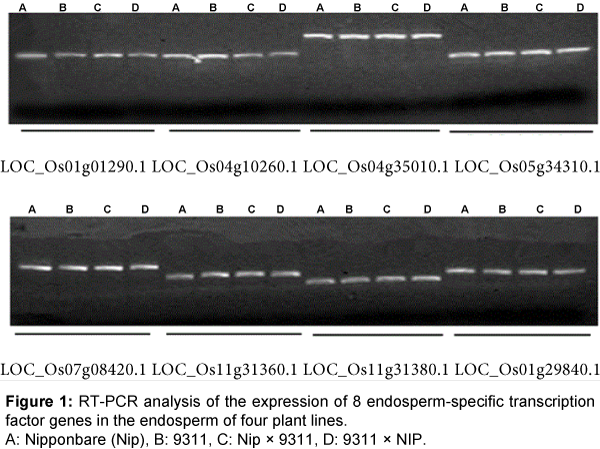
Based on the endosperm-specific genes identified by Zemach [38], 26 genes encoding transcription factors were chosen in our analysis. We investigated whether there were SNPs in these genes in Japonica rice Nip and non-glutinous rice 9311 based on information obtained from the PubMed website. The gene regions that are different compared with parents were used to design PCR primers, and endosperm cDNA (shown in experimental materials) was used as a template for PCR. A total of 18 genes could be amplified by PCR and RT-PCR results of 8 genes are shown in Figure 1.
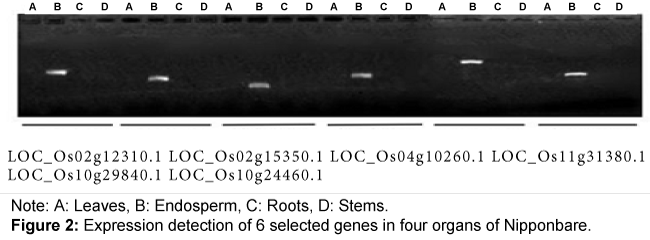
To confirm if these 18 genes are specifically expressed in endosperm, we synthesized cDNA using PCR amplification from RNA that were isolated from leaves, endosperm, roots, and stems of Nip. All 18 genes could only be amplified from endosperm tissue but not from leaf, root, or stem tissue, demonstrating that these 18 genes were specifically expressed in endosperm (Figure 2).
Identification of non-imprinted genes
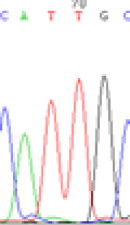
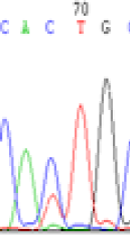
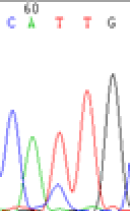
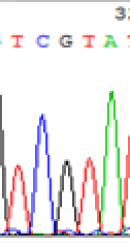
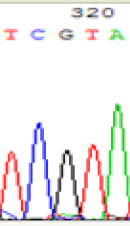
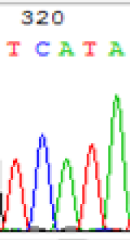
We used Sanger sequencing to test the imprinting status of selected endosperm specific genes. The amplified bands were sequenced to examine the expression of SNPs loci containing the parental differences in the progeny plants. If the differences in the expression of SNPs of the parents simultaneously expressed in the progeny, indicating that the gene was non-imprinted gene; conversely, if the progeny combinations only expressed one of the parental different bases, then the gene was imprinted gene. The sequencing results found that among 18 genes, there were 4 non-imprinted genes and 14 imprinted genes. The Sanger sequencing results of 6 genes (4 non-imprinted genes, 2 imprinted genes) were shown in in Table 2.
| Combination | Nip | 9311 | Nip × 9311 | 9311 × Nip |
|---|---|---|---|---|
| SNPSSNPs | CACTG | CATTG | CAC(T)TG | CAT(C)TG |
| LOC_Os04g35010.1(non-imprinted) | 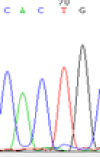 |
 |
 |
 |
| SNPSSNPs | GACAT | GATAT | GAC(T)AT | GAT(C)AT |
| LOC_Os07g08420(non-imprinted) | 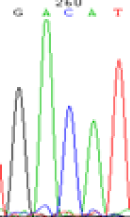 |
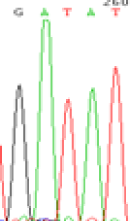 |
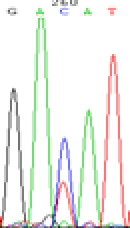 |
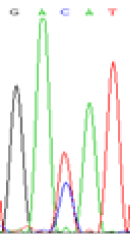 |
| SNPSSNPs | TGCCA | TGTCA | TGC(T)CA | TGT(C)CA |
| LOC_Os02g12310.1(non-imprinted) | 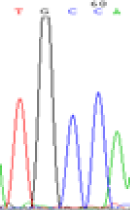 |
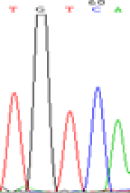 |
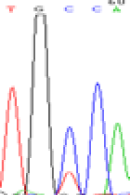 |
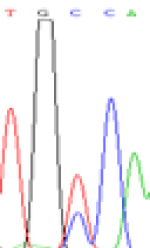 |
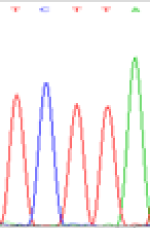
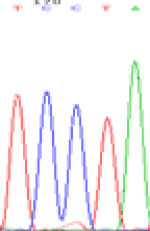
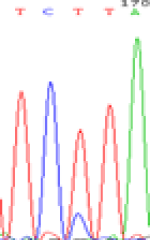
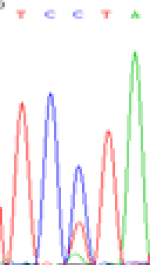
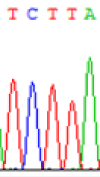
| SNPSSNPs | TCTTA | TCCTA | TCT(C)TA | TCC(T)TA |
| LOC_Os01g24460.1(non-imprinted) | 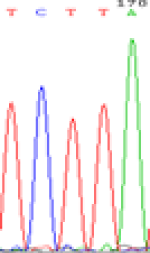 |
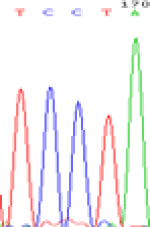 |
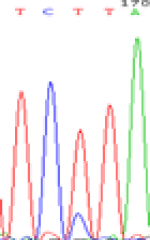 |
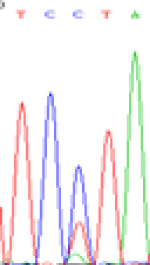 |
| TCATA | TCGTA | TCGTA | TCATA | |
| LOC_Os01g33350.1(imprinted) | 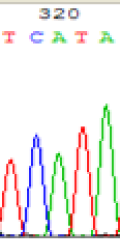 |
 |
 |
 |
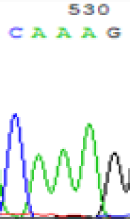
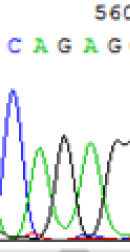
| SNPs | CAGAG | CAAAG | CAGAG | CAAAG |
| LOC_Os10g25850.1(imprinted) | 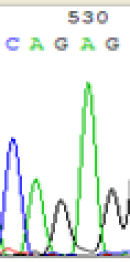 |
 |
 |
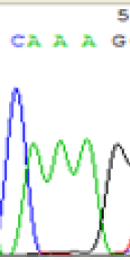 |
Table 2: Comparison of SNP expression of endosperm-specific genes in parents and hybrid seedlings.
The effect of 5-azadC-mediated demethylation on the expression of endosperm specific genes in hybrid seedlings
DNA methylation regulates the expression of imprinted genes in the endosperm, but it is not yet known whether methylation is also involved in regulating the expression of non-imprinted genes. Therefore, we treated rice seeds with the methylation inhibitor 5-azadC (30 μmol/L) for 3-5 d, followed by germination, to examine the effects of methylation on the regulation of non-imprinted genes.
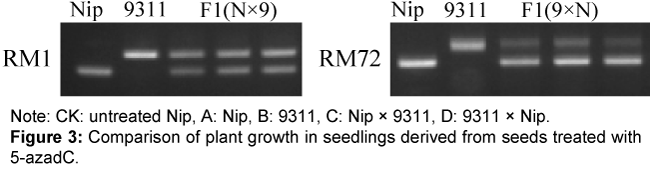
We first examined the true and false hybrid plants and to ensure the authenticity of the hybrids, we used 38 pairs of microsatellite primers to differentiate between genes from Nipponbare and 9311 and obtained 11 different markers (RM1, RM4, RM9, RM18, RM47, RM52, RM72, RM335, RM337, RM339, and RM341). Markers RM1 and RM72 were used to confirm the identity of the hybrids by examining the leaves of the F1 hybrid plants, as both markers should be amplified simultaneously in true hybrids. The results confirm that the hybrids utilized in this study were true hybrids (Figure 3).
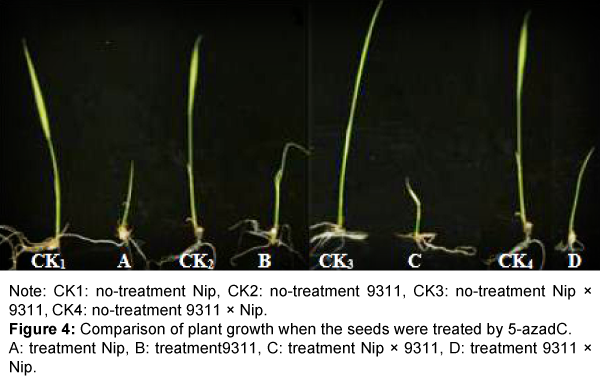
After inhibitor treatment, the5-azadC-treated seeds exhibited a delayed germination and the plants became stunted compared with the control (Figure 4). The latter phenomenon was changed in most plants after 5 days of growth except some plants wilted and gradually died. These results are consistent with the findings of Sano et al. [41].
The expression of the 18 endosperm-specific genes in the leaves of plants produced from 5-azadC-treated seeds were investigated using RT-PCR. It turned out that only four genes could be amplified from these leaves (Figure 5), indicating that the expression of these four genes is regulated by DNA methylation, i.e., de-methylation enabled these genes to be expressed in leaves.
Activation of various parental alleles by demethylation in hybrid seedlings
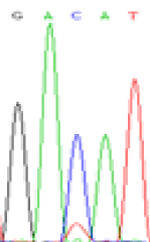
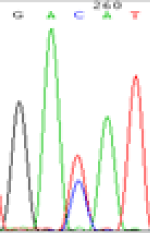
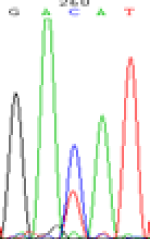
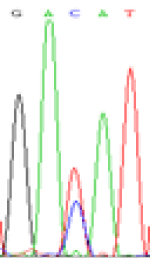
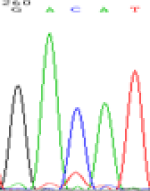
RNA was extracted from the leaves of seedlings derived from 5-azadC-treated seeds and subjected to RT-PCR to investigate the expression of parental alleles at the four non-imprinted loci aforementioned and to see whether the expression patterns of SNP loci in leaves were consistent with those in endosperm. In the endosperm of hybrids, gene LOC_Os07g08420 expressed the SNPs C and T simultaneously that come from mother and father, respectively. In the seeds treated with 5-azadC, the new leaves (demethylated leaves) still expressed the bases C and T, suggesting that 5-azadC treatment did not influence the expression of parental alleles of this gene in the treated hybrid seedlings. As LOC_Os 07g08420 is only expressed in endosperm and silenced in embryos and other tissues. We speculate that DNA methylation plays an essential role in silencing the activity of this gene in non-endosperm tissues. On the same line, we observed that seeds treated with 5-azadC showed the expression of both parental alleles in the seedlings at this locus (Table 3).
| Endosperm-3d | Endosperm-5d | 5-azadC–leaf | |||
|---|---|---|---|---|---|
| Nip × 9311 | 9311 × Nip | Nip × 9311 | 9311 × Nip | Nip × 9311 | 9311 × Nip |
| GAC(T)AT | GAT(C)AT | GAC(T)AT | GAT(C)AT | GAC(T)AT | GAC(T)AT |
 |
 |
 |
 |
 |
 |
Table 3: The expression of C and T SNPs at LOC_Os07g08420 in two developmental stages of endosperm and the demethylated leaves after 5-azadC treatment.
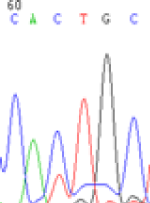
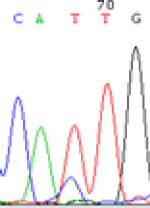
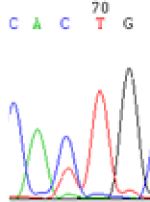
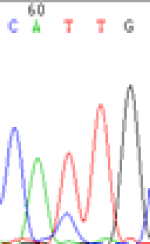
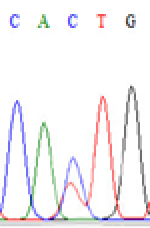
Interestingly, gene LOC_Os04g35010.1 was similarly activated like LOC_Os 07g08420 (Table 4) thus suggesting that DNA methylation is involved in regulating the expression of endosperm specific nonimprinted genes.
| Endosperm-3d | Endosperm-5d | AzadC-leaf | |||
|---|---|---|---|---|---|
| Nip × 9311 | 9311 × Nip | Nip × 9311 | 9311 × Nip | Nip × 9311 | 9311 × Nip |
| CAC(T)TG | CAT(C)TG | CAC(T)TG | CAT(C)TG | CAC(T)TG | CAC(T)TG |
 |
 |
 |
 |
 |
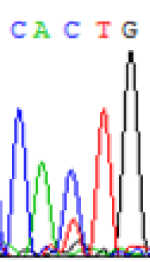 |
Table 4: The expression of C and T SNPs at LOC_Os04g35010.1 in two development stageal s of endosperm and the demethylationed leaves after 5-azadC treatment.
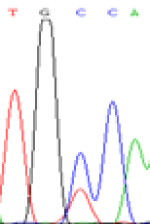
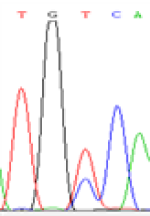
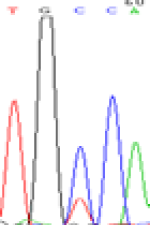
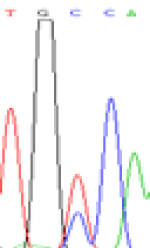
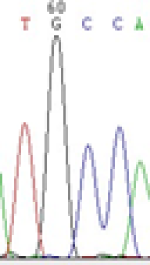
Furthermore, gene LOC_Os02g12310 expressed the polymorphism loci C and T in the 3d and 5d endosperm simultaneously as we expected. However, we observed that only the Nip allele (C) was activated after 5-azadC treatment while the 9311 remains silenced. Obviously there are differential effects of DNA methylation on the Nip and 9311 alleles. But it is not clear why 9311 is not activated (Table 5).
| Endosperm-3d | Endosperm-5d | AzadC-leaf | |||
|---|---|---|---|---|---|
| Nip × 9311 | 9311 × Nip | Nip × 9311 | 9311 × Nip | Nip × 9311 | 9311 × Nip |
| TGC(T)CA | TGT(C)CA | TGC(T)CA | TGT(C)CA | TGCCA | TGCCA |
 |
 |
 |
 |
 |
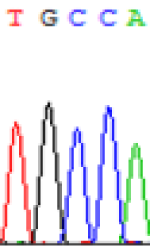 |
Table 5: The expression of C and T SNPs at LOC_Os02g12310 in two developmental stages of endosperm and the seedlings after 5-azadC treatment.
Similarly, LOC_Os01g24460 expressed the SNPs C and T in the 5d endosperm simultaneously (Table 6). Interestingly at 3d only the maternal allele is detected in the endosperm, suggesting that the imprinting process could be dynamic (Table 6). Strikingly the hybrid seedlings with reduced DNA methylation after inhibitor treatment showed the same maternal expression at this locus as shown in the endosperm of 3d. But the underlying mechanisms driving the imprinted expression in early endosperm and treated seedlings remain to be determined.
| Endosperm-3d | Endosperm-5d | AzadC-leaf | |||
|---|---|---|---|---|---|
| Nip × 9311 | 9311 × Nip | Nip × 9311 | 9311 × Nip | Nip × 9311 | 9311 × Nip |
| TCTTA | TCCTA | TCT(C)TA | TCC(T)TA | TCTTA | TCCTA |
 |
 |
 |
 |
 |
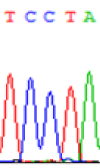 |
Table 6: The expression of C and T bases SNPs at LOC_Os01g24460.1 in Endosperm two developmental stages of e and the leaves seedlings after 5-azadC treatment.
Together, expression patterns at LOC_Os01g24460 are related to development stages, namely imprinted expression occurs in the 3d endosperm, but not in the 5d endosperm. This dynamic process may require the conversion of imprinted locus into non-imprint one at later development stage and DNA de-methylation may be involved in this process. Our results also showed that DNA methylation is also involved in the regulation of the selective demethylation of the maternal alleles of the seedlings being treated by 5-azadC at this locus.
Discussion
DNA methylation regulates on endosperm specific genes and may repress the activity of deleterious gene in seedlings
The expression levels of many genes are dynamic in different organs and at different developmental stages. The mechanisms regulating the spatial and temporal expression of genes include both epigenetic modifications and the role of regulatory factors. It is known that methylation inhibitor 5-azadC can reduce DNA methylation levels, thereby leading to the change of gene expression [39-42]. For endosperm-specific genes, we have tested whether the silencing in non-endosperm tissues are due to DNA methylation. We detected the expression of endosperm-specific genes in the seedlings derived from rice seeds treated with 5-azadC. Further sequencing analysis of the seedling-expressed genes showed that there were two types of expression patterns: either both parental alleles as represented by polymorphic loci (SNPs) or only one of the polymorphic loci (SNPs) were expressed in the leaves derived from inhibitor treated seeds.
According to functional annotation [38], LOC_Os07g08420 plays an important role in grain filling of rice, LOC_Os04g35010.1 encodes DNA helix domain protein, LOC_Os02g12310 encodes no apical meristem protein (NAC family), and LOC_Os01g24460.1 encodes histone-like transcription factors. Seeds treated with 5-azadC showed delayed germination, dwarfing of plant height, death at late development stage and other detrimental phenotypes. It remains to be determined whether activation of the endosperm-specific genes of diverse functions contributes to the detrimental phenotypes after seed treatment. It is possible that demethylation mediated by 5-azadC would activate many more genes that would be silenced in seedlings and, most likely these ectopic activations will cause abnormal plant development. Consistently, it is hypothesized that evolutionary selection of imprinted genes (which usually are endosperm specific and silenced by DNA methylation) is driven by silencing deleterious gene activity in somatic tissues [3]. Interestingly, LOC_Os01g24460 showed imprinted expression in early endosperm development.
It has been shown DNA methylation patterns and the changes in its expression levels were closely related to the formation of heterosis [28-33]. Chodavarapu et al. [30] found that the difference of cytosine methylation level between rice parents (Nip and 9311) and F1 was 0.79%, thus they hypothesized that the heterosis of F1 was related to the reduction of DNA methylation levels. Zhiben et al. [28-29,31-32] also found that DNA methylation had a close relationship to heterosis in rice, maize and sorghum, respectively. At the LOC_Os01g24460 locus only the Nip allele was detected in the hybrid seedlings after removal of DNA methylation, suggesting that the Nip locus had a heterosis in the progeny. The other three genes also showed activation of parental alleles in F1 hybrid derived from treated seeds.
LOC_Os01g24460 in the 3d endosperm displayed maternal expression, but the expression at 5d is bi-allelic, indicating that imprinting is a dynamic process. Strikingly, after removing DNA methylation, LOC_Os01g24460 also expressed as a maternal imprinting gene in the hybrid leaves, suggesting that the maternal alleles of this gene in both endosperm and 5-azadC treated seedlings are prone to demethylation process that drives imprinted expression. What causes the differential responses to 5-azadC mediated demethylation process in hybrid seedlings is unknown. In contrast, DEMETER mediated demethylation process at the maternal alleles in endosperm has been clearly demonstrated in various plants.
Du [42] also found that an imprinted gene yellow2-like in the seedling derived from rice seeds treated with DNA methylation inhibitor 5-azadC or zebularine could produce novel transcripts, indicating that DNA methylation can inhibit the transcription initiation of abnormal promoters in plants, thereby resulting in specific expression. This finding together with our study provide evidence that DNA methylation at least is one of the main mechanisms that drive tissue-specific gene expression and DNA methylation is important for plant development.
DNA methylation is associated with Heterosis and imprinting
In angiosperms, seed development and germination are related processes because embryo development and germination require nutriment flow from endosperm. Most imprinted expression is observed in the endosperm of developing seed. Previous studies suggest that the transposon insertion that affect the expression of neighboring genes is the main driving force of the evolution of genomic imprinting [25-27,43]. DNA methylation is considered to silence the deleterious insertion of transposons. Therefore, large numbers of imprinted genes are endosperm specific because these genes are silenced in non-endosperm tissues due to DNA methylation and activated by DEMETER that remove DNA methylation. For nonimprinted endosperm-specific genes, their role and the regulation of their expression are largely unknown. Current study analyzed the role of DNA methylation modification in the regulation of non-imprinted genes and detected expression of the parental SNPs of four nonimprinted genes in demethylated hybrid seedlings. Our results support the idea that there is a close relationship between the dynamic changes in DNA methylation and the establishment of imprinting as well as, potentially, heterosis.
In this study we have only selected 4 endosperm-specific transcription factors to analyze the role of DNA methylation in the regulation of gene expression, it is still required to perform s more indepth analysis on the regulation of other transcription factors and nontranscription factors that are expressed specifically in endosperm. It is known that DNA methylation, histone H3K27 modification, POIIVdependent siRNAs (p4-siRNAs) and other epigenetic mechanisms are involved in the expression of imprinted genes and the formation of heterosis [25,33].
References
- Jullien PE, Katz A, Oliva M, Ohad N, Berger F (2006) Polycomb group complexes self-regulate imprinting of the polycomb group gene MEDEA in Arabidopsis. CurrBiol 16: 486-492.
- Huh JH, Bauer MJ, Hsieh TF, Fischer R (2007) Endosperm gene imprinting and seed development. CurrOpin Genet Dev 17: 480-485.
- Berger F, Chaudhury A (2009) Parental memones shape seeds. Trends Plant Sci 14: 550-556.
- Bauer MJ, Fischer RL (2011) Genome demethylation and imprinting in the endosperm. CurrOpin Plant Biol 14: 162-167.
- Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, (2009) Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451-1454.
- Haun WJ, Springer NM (2008) Maternal and paternal alleles exhibit differential histone methylation and acetylation at maize imprinted genes. Plant J 56: 903-912.
- Gutierrez-Marcos JF, Costa LM, Pra DM, Scholten S, Kranz E, et al. (2006) Epigenetic asymmetry of imprinted genes in plant gametes. Nat Genet 38: 876-878.
- Haig D, Wilczek A (2006) Sexual conflict and the alternation of haploid and diploid generations. Philos Trans R SocLond B BiolSci 361: 335-343.
- Dilkes BP, Comai L (2004) A differential dosage hypothesis for parental effects in seed development. Plant Cell 16: 3174-3180.
- Babak T, Deveale B, Armour C, Raymond C, Cleary MA, et al. (2008) Global survey of genomic imprinting by transcriptome sequencing. CurrBiol 18: 1735-1741.
- Hagan JP, O'Neill BL, Stewart CL, Kozlov SV, Croce CM (2009) At least ten genes define the imprinted Dlk1-Dio3 cluster on mouse chromosome 12qF1. PLoS One 4: e4352
- Arnaud P, Feil R (2006) MEDEA takes control of its own imprinting. Cell 124: 468-470.
- Stoute AI, Varenko V, King CJ, Scott RJ, Kurup S (2012) Parental genome imbalance in brassica oleracea causes asymmetric triploid block. Plant J 71: 503-516.
- Scott RJ, Spielman M, Bailey J, Dickinson HG (1998) Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125: 3329-334.
- Rapp RA, Udall JA, Wendel JF (2009) Genomic expression dominance in allopolyploids. BMC Biology 7: 18.
- Sekine D, Ohnishi T, Furuumi H, Ono A, Yamada T, et al. (2013) Dissection of two major components of the post-zygotic hybridization barrier in rice endosperm. Plant J 76: 792002D799
- Kirkbride RC, Yu HH, Nah G, Zhang C, Shi X, et al. (2015) An epigenetic role for disrupted paternal gene expression in postzygotic seed abortion in Arabidopsis interspecific hybrids. Mol Plant 8: 1766-1775.
- Jullien PE, Berger F (2010) Parental genome dosage imbalance deregulates imprinting in Arabidopsis. PLoS Genet 6: p.e1000885.
- Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, et al. (2002) DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110: 33-42.
- Xiao W, Gehring M, Choi Y, Margossian L, Pu H, et al. (2003) Imprinting of the MEA polycomb gene is controlled by antagonism between METlmethyltransferase and DME glycosylase. Dev Cell 5: 891-901.
- Kinoshita T, Miura A, Choi Y, Klnoshlta Y Cao X, et al. (2004) One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303: 521-523.
- Zhang WW, Cao SX, Jiang L, Zhu SS, Wan JM (2005) Genomic imprinting and seed development. Genetics 27: 665-670.
- Luo M, Taylor JM, Spriggs A, Zhang HY, Wu XJ, et al. (2011) A genome-wide survey of impnnted genes in rice seeds reveals imprinting pdmadly occurs in the endosperm. PLoS Genet7.e1002125
- Hsieh TF, Shin J, Uzawa R, Silva P, Cohen S, et al. (2011) Regulation of imprinted gene expression in Arabidopsis endosperm. ProcNatlAcadSci U S A 108: 1755-1762.
- Zhang M, Liu B (2012) Epigenetic regulation of endosperm development in plant. ActaBOtanicaSinica 47: 101-110.
- Zhang H, Chaudhury A, Wu X (2013) Imprinting in Plants and Its Underlying Mechanisms. J Genet Genomics 40: 239-247
- Zhang Hongyu, XuPeizhou, Yang Hua (2010) Emprinted genes in Arabidopsis and expression regulation of imprinting. Genetics 32: 670-676.
- Yi Zhiben, Sun Yi, NiuTiantang, et al. (2005) Study on difference of cytosine methylation of sorghum genome DNA between hybrids and parents. Crop Sinica 31: 1138-1143.
- Hepburn PA, Margison GP, Tisdale MJ (1991) Enzymatic methylation of cytosine in DNA is prevented by adjacent O6-methylguanine residues. J BiolChem 266: 7985-7987
- Chodavarapu RK, Feng S, Ding B, Simon SA, Lopez D, et al. (2012) Transcriptome and methylome interactions in rice hybrids. ProcNatlAcadSci USA 109: 12040-12045.
- Tsaftaris AS, Kafka M (1998) Mechanism of heterosis in crop plants. J Crop Prod 1: 95-111.
- Xiong LZ, Xu CG, SaghaiMaroof MA, Zhang Q (1999) Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol Gene Genet 261: 439-446.
- Cui Huihui, Xiang Chao, Shi Yingyao, Wang Wensheng, GaoYongming, et al. (2015) Research progress in epigenetics of heterosis. Journal of Plant Genetic Resources 16: 933-939.
- Li Na, Zhang Yang, XieLinan (2012) Research progress in plant DNA methylation. Plant Physiology Journal 48: 1027-1036.
- GuoGuangping, Yuan Jinling, Wu Xiaoli (2011) Application status and prospects of DNA methylation in plant research. Journal of Plant Genetic Resources 12: 425-430.
- Jin H, Hu W, Wei Z, Wan L, Li G, et al. (2008) Alterations in cytosine methylation and species-specific transcription induced by interspecific hybridization hybridization between Oryza sativa and O. Officinalis. TheorAppl Genet 117: 1271-1279.
- Zhao Y, Yu S, Xing C, Fan S, Song M (2008) Analysis of DNA methylation in cotton hybrids and their parents. MolBiol (Mosk) 42: 195-205.
- Zemach A, Kim MY, Silva P, Rodrigues JA, Doston P, et al. (2010) Local DNA hypomethylation activates genes in rice endosperm.ProcNatlAcadSci USA 107: 18729-18734.
- Wang J, Tian L, Madlung A, Lee HS, Chen M, et al. (2004) Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 167: 1961-1973.
- Lee HS, Chen ZJ (2001) Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. ProcNatlAcadSci USA 98: 6753-6758
- Sano H, Kamada I, Youssefian S, Katsumi M, Wabiko H (1990) A single treatment of rice seedlings with 5-azacytidine induces heritable dwarfism and undermethylation of genomic DNA. Molecular and General Genetics MGG 220: 441-447
- Du M, Luo M, Zhang R, Finnegan EJ, Koltunow AM (2014) Imprinting in rice: the role of DNA and histone methylation in modulating parent-of-origin specific expression and determining transcript start sites. Plant J 79: 232-242.
- Ishikawa R, Kinoshita T (2009) Epigenetic programming: The Challenge to Species Hybridization. Mol Plant 2: 589-599.
Relevant Topics
- Basmati Rice
- Drought Tolerence
- Golden Rice
- Leaf Diseases
- Long Grain Rice
- Par Boiled Rice
- Raw Rice
- Rice
- Rice and Aquaculture
- Rice and Nutrition
- Rice Blast
- Rice Bran
- Rice Diseases
- Rice Economics
- Rice Genome
- Rice husk
- Rice production
- Rice research
- Rice Yield
- Sticky Rice
- Stress Resistant Rice
- Unpolished Rice
- White Rice
Recommended Journals
Article Tools
Article Usage
- Total views: 12082
- [From(publication date):
May-2016 - Apr 05, 2025] - Breakdown by view type
- HTML page views : 11139
- PDF downloads : 943