Research Article Open Access
The Burden of HIV Associated Drug Resistance Mutations in an Early Infant Diagnosis Program: A Glance through the Paediatric Window of Zimbabwe
| Monalisa T Manhanzva1*, Junior Mutsvangwa2, Ingrid A Beck3, Lisa M Frenkel3,4, Mqondisi Tshabalala5, Justen Manasa6, Alltalents T Murahwa1,5 and Lovemore Gwanzura1 | ||
| 1University of Zimbabwe College of Health Sciences, Department of Medical laboratory Sciences, P.O. Box A178 Avondale, Harare, Zimbabwe | ||
| 2Biomedical Research and Training Institute, Harare, Zimbabwe | ||
| 3 University of Washington, Seattle WA USA | ||
| 4Seattle Children's Research Institute, Seattle, 1900 9th Avenue, Washington, USA | ||
| 5University of Zimbabwe College of Health Sciences, Department of Immunology, P.O. Box A178 Avondale, Harare | ||
| 6African Centre for Health and Population Studies, University of KwaZulu-Natal, Durban, South Africa | ||
| Corresponding Author : | Monalisa T. Manhanzva University of Zimbabwe College of Health Sciences Department of Medical laboratory Sciences P.O. Box A178 Avondale, Harare, Zimbabwe E-mail: monalisatatenda@gmail.com |
|
| Received October 25, 2014; Accepted January 03, 2014; Published January 07, 2014 | ||
| Citation: Manhanzva MT, Mutsvangwa J, Beck IA, Frenkel LM, Tshabalala M, et al. (2015) The Burden of HIV Associated Drug Resistance Mutations in an Early Infant Diagnosis Program: A Glance through the Paediatric Window of Zimbabwe. J Infect Dis Ther 3:198. doi:10.4172/2332-0877.1000198 | ||
| Copyright: ©2015 Manhanzva MT, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | ||
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
||
Visit for more related articles at Journal of Infectious Diseases & Therapy
Abstract
There is paucity of information on the prevalence of human immunodeficiency virus type 1 drug-resistance mutations in infants infected despite prevention of mother to child transmission in Zimbabwe. This study examined 32 dried blood spot specimens from HIV- 1 C infected infants born alive to women receiving antiretroviral therapy, who were part of the WHO/ResNet early infant diagnosis program between January 2010 and January 2011. The objective was to determine the patterns and levels of HIV drug resistance and inform on policy formulation. Overall HIV prevalence of the infants in the retrospect study period was 2.4% (32/1356). Half of the samples (16/32) analyzed had HIV associated drug resistance mutations, however excluding polymorphic mutations the HIV drug resistant mutations were 25% (8/32). Frequencies of the mutations were (E138A, n=6; G190A, n=1; M230ML, n=1; K103KN, n=1; Y181C, n=5; V90VI, n=1; E138G, n=1; E138EA, n=1). One patient had two mutations the K103KN and the Y181C. Of the sixteen patients with HIV drug resistance mutations, 5 had nevirapine only resistance, none had lamivudine resistance and 8 had both etravirine and rilpivirine resistance, 2 had both nevirapine, efavirenz and 1 had rilpivirine only. The study shows an apparent non-nucleoside reverse transcriptase inhibitor (NNRTI) drug resistance predominance and suggests a judicious use of NNRTI regimens and a prudent strategy to minimize the selection of drug resistance mutations.
| Keywords | |
| Mutations; HIV; Pregnancy; Sequencing | |
| Introduction | |
| UNAIDS estimated 35.3 (32.2-38.8) million people to be living with HIV infection by 2012, of which 3.3 (3.0-3.7) million and 17.7 (16.4-19.3) million were children under 15 years of age and women respectively. Zimbabwe (adult prevalence 14.7 %), is part of sub Saharan Africa with the highest global (69%) HIV burden [1]. It has also been estimated that 91% of all pregnant women living with HIV in low and middle income countries live in 25 countries, about 70% live in 10 countries which include Kenya, Mozambique, Nigeria, South Africa and Zimbabwe [2]. | |
| The burden of the HIV epidemic in children reflects the risk factors for HIV infection in the adult population, the frequency of undetected maternal HIV infection and the magnitude of mother –to- child transmission (MTCT) [3]. HIV infection through MTCT accounts for more than 90% of HIV infections amongst children, hence the critical need to identify HIV infected pregnant and lactating mothers in order to manage them appropriately to reduce transmission rates [4]. | |
| As a recommendation by the world health organization (WHO), infants who are known to have been exposed to HIV should be tested by a polymerase chain reaction (PCR) at 4-6 weeks of age using dried blood spots (DBS) [5] to reduce the pandemic in children. | |
| The high burden of the HIV pandemic in sub Sahara is further confounded by the interlinked issues of poverty and limited access to health facilities, resulting in an increased risk of MTCT of HIV [6]. This justifies the urgency of the introduction of interventions to reduce MTCT. In addition HIV treatment in children is more challenging compared to adults’ treatment strategies; with a higher risk of virological failures among children [3]. Other challenges in treating HIV infected infants include limited pediatric formulations [7-9] and a myriad of challenges like pediatric HIV diagnosis and lack of resources [10-13]. | |
| Without any intervention, 15-45% of babies born to HIV infected mothers acquire the virus during pregnancy, labor and breast feeding, with about 65 % of these infant HIV cases likely to occur peripartum [14,15]. With the advent of highly active antiretroviral therapy (HAART), several regimens have been recommended for prevention of mother to child transmission (pMTCT) with varying outcomes [16-18]. Since MTCT is the primary means by which children become HIV infected, strategies to reduce the transmission rates have focused on the time period during pregnancy, intrapartum, post partum and during breastfeeding [19]. | |
| The WHO guidelines as of June 2013 recommend that either antiretroviral (ARV) drugs be given to women living with HIV during pregnancy and breast feeding or lifelong antiretroviral therapy for all pregnant and breastfeeding women who are living with HIV. The former being option B and the latter option B + [20]. Option A regimen, which is being phased out consisted of maternal zidovudine (AZT) plus infant ARV prophylaxis i.e. ante partum twice daily AZT, plus single dose nevirapine (sd-NVP) at the onset of labour, plus twice daily AZT + lamivudine (3TC) during labour and delivery and continued for seven days post-partum [2]. | |
| In Zimbabwe, antiretroviral therapy for pMTCT of HIV has been available since 2003, with sd-NVP being the regimen of choice (by then) in which the mother was given a dose of nevirapine at the onset of labor i.e. at least 2 hours before delivery and another dose was administered to the infant within 72 hours of delivery [21]. Studies have shown that a single dose of sd-NVP reduces MTCT by approximately 47% [22] making it ideal for resource poor settings like Zimbabwe due to its economic feasibility and simplicity [6]. However, sd-NVP has been associated with the emergence of drug resistance in the exposed individuals and hence the regimen was phased out of the pMTCT programmes [17]. The resistance mutations that have been associated with NVP resistance include K103N, V106M, Y181C and G190A [23]. HIV drug resistance mutations associated with sd-NVP use pose a negative impact on strides made in pMTCT and HIV treatment in general especially in resource limited [24,25]. The scale– up of ART is based on using two nucleoside reverse transcriptase inhibitors (NRTIs) and one non-nucleoside reverse transcriptase inhibitor (NNRTI). In low to middle income countries, this has gone a long way in reducing AIDS related morbidity and mortality and MTCT of HIV [26]. However, increased and injudicious use of ARVs for paediatric HIV prevention in pregnant women will lead to an increase in the number of HIV infected children despite pMTCT as a result of drug resistance [27-31]. | |
| In this study, the authors hypothesized emergence of HIV drug resistance mutations in children exposed to 3TC and possibly AZT which are being given as part of the most efficacious regimen, option B and option B+ respectively in Zimbabwe following abandonment of sd-NVP. The hypothesis is based on emergence of HIV drug resistance mutations in children exposed to ART during the use of sd-NVP in pMTCT programmes. This data will elaborate the extent of HIV-1 drug resistance in children in Zimbabwe with the view of improving pMTCT strategies and reducing the HIV burden. Zimbabwe is part of the early infant diagnosis program (EID), a WHO, HIVResNet and centers for disease control (CDC) prevention and surveillance initiative to assess HIV drug resistance (HIVDR) to specific ARVs among infants aged < 18months of age and newly infected with HIV in resource poor settings [5]. | |
| This study sought to document HIV drug resistance mutations in a cohort of HIV-1 C infected infants born to mothers that were in the Option A pMTCT program in Zimbabwe. We thus report here the prevalence of HIVDR mutations in HIV-1 C infected infants born alive to women receiving ART for their health and as infant prevention with twice daily AZT ante-partum, sd-NVP at labour onset and 3TC continued for one week. | |
| Methodology | |
| Ethical approval and considerations | |
| Ethical approval was obtained from the Joint Parirenyatwa and College of Health Sciences Research Ethics Committee (JREC) and the national Medical Research Council of Zimbabwe (MRCZ) with the reference numbers JREC/127/11 and MRCZ/B/228 respectively. No procedures or interventions were done directly to the participants: the study was done using archived DBS samples that had already been collected for EID of HIV. The study adhered to the Helsinki declaration. | |
| Specimen selection and sample size | |
| Specimens were obtained from the archives of the National Microbiology Reference Laboratory (NMRL) of Zimbabwe as part of the ongoing EID study focusing on HIV diagnosis among children <18 months of age. Permission to access specimens was granted by the EID National Study coordinator together with ethical approval as outlined above. Thirty two (32) DBS samples of all HIV-1 positive infants (<6 months) born alive to women receiving 3TC and NVP containing regimens (obtained between January 2010 and January 2011) were retrieved for HIV DR analysis. | |
| DNA Extraction from the DBS samples | |
| DNA extraction from the 32 frozen (-80°C) DBS samples was done using the DNA Extraction from Filter Paper with Chelex-100 protocol described elsewhere [32]. In this procedure, 50 μl spots were excised from each blood saturated DBS using quarter punches and placed into 1.5 ml screw capped tubes. The punchers were cleaned by punching a clean filter paper 4-5 times in-between each patient blood spot according to protocol. A wash buffer constituting of phosphate buffered saline (PBS) and triton X-100 was used to wash off the blood from the filter paper by vortexing the mixture briefly and then incubated at room temperature for 15 minutes on a shaker. After this, the samples were centrifuged at 13 000rpm and the supernatant was discarded. The wash was repeated once and then 200 μl of 10% chelex was added to the tube containing the washed filter paper. The tubes were placed on a heat block and incubated at 56áµï¿½C for 1½ hrs. The tubes were removed from the heat block and centrifuged, then placed into a boiling water bath for 10 minutes; the supernatant containing the DNA was carefully transferred to labeled clean 1.5 ml screw capped tubes and stored at -20áµï¿½C. | |
| PCR Amplification | |
| The HIV-1 DNA from the 32 DBS samples was amplified by nested PCR using primers specific for the HIV-1 reverse transcriptase gene codons 1 to 240 using a PTC–100 Peltier thermal Cycler as previously described [32]. Briefly; two sets of primers are used sequentially, with the first primer set binding to sequences outside the target DNA in the first round. The second primer set binds to sequences in the target DNA that are within the portion amplified by the first set in the second round. The primer sets for PCR were MozFO: 5’CCTACACCTGTCAACATAATTGG3’forward primer and NER10: 5’AAYTTCTGTATATCATTGACAGTCCA3’ reverse primer, and. MozFI: 5’AATTAAAGCCAGGAATGGATGG3’ forward primer and NER11: 5’CAYTTGTCAGGATGGAGTTCATA3’ reverse primer for first and second round PCR respectively. The first round PCR reaction (80 μl reaction volume) tube contained of PCR buffer, 2 mM dNTPs, MgCl2, nuclease free water, the enzyme taq polymerase, the forward primers Moz F0, the reverse primers NER10 and 20ul of DNA extract. Second round PCR reaction tube had a reaction volume of 50ul containing PCR buffer, 2 mM dNTPs, taq DNA polymerase, forward primer MozF1 and reverse primers NER11. The two rounds were critical to obtain enough PCR products for subsequent analysis. The cycling conditions consisted of a denaturation of 94áµï¿½C for 5minutes followed by 35 cycles of 94áµï¿½C for 30 seconds, 55áµï¿½C for 30 seconds, an extension of 72áµï¿½C for 1minute and a final extension at 72áµï¿½C for 7 minutes. The cycling conditions for the second round PCR were similar to those for the first round PCR. After the second round PCR, a 684 base pair DNA fragment, corresponding to the amplified HIV-1 C product was assessed by ethidium bromide stained 1% agarose gel electrophoresis. A positive control containing 10 copies of HIV-1 C was included in each PCR reaction. Negative controls containing all the PCR reagents excluding template DNA were also included in every PCR run to ensure there was no contamination of any of the reagents. | |
| HIV reverse transcriptase sequencing | |
| Samples that gave the characteristic 684bp band were sequenced by the dye terminator method as described [33], briefly; PCR amplicons were cleaned up using the PureLink QUICK PCR Purification Kit (Life Technologies, Foster City, CA) and sequenced using the Big Dye Terminator kit version 3.1 (Life Technologies, Foster City) and a set of bidirectional primers. Four separate sequencing reactions with each of the four primers were prepared for each sample. Ten microliter reactions with 6.1 μl water, 2 μl 5X Sequencing buffer (Life Technologies), 0.5 μl 3.2 pmol/ml of sequencing primers, 0.4 μl of the Big Dye Terminator Sequencing mix and 1 μl of the cleaned PCR product were used for sequencing. The cycling condition for the sequencing were 35 cycles of 96°C for 10 seconds, 50°C for 5 seconds and 60°C for 4 minutes followed by a hold a 4°C. The ethanol-sodium acetate protocol was used for the clean-up of the sequencing products. The cleaned sequencing PCR products were re-constituted in 10 μl of HiDi-formamide and denatured at 95°C for 3 minutes. Capillary sequencing electrophoresis was done on a 3130XL genetic analyzer (Life Technologies, Foster City, CA). | |
| Sequencing analysis | |
| The sequences covering the first 240 codons of the reverse transcriptase gene were assembled using Geneious Pro genetic analyzer [34]. The quality of the sequences was assessed using the HIV-1 Quality Analysis Tool [35] and the Calibrated Population Resistance (CPR) tool [36]. HIV-1 subtyping was performed using the REGA HIV-1 Subtyping Tool v 2.0 [37]. Phylogenetic analysis was done to aid with quality assurance of the sequencing. | |
| Statistical analysis | |
| Stata version 10 (StataCorp LP, Texas, USA) was used for analyzing the data. Baseline description of study participants was presented using percentages for categorical variables such as gender. Prevalence of HIV drug resistance mutations was calculated as percentages. | |
| Results | |
| Demographic characteristics | |
| Of the 32 DBS specimens included in the study 47% (15) were from female and 53% (17) male infants. The age ranges of the infants from whom the specimens were obtained ranged from 0-6months as per the national EID database. | |
| Prevalence of HIV drug resistance | |
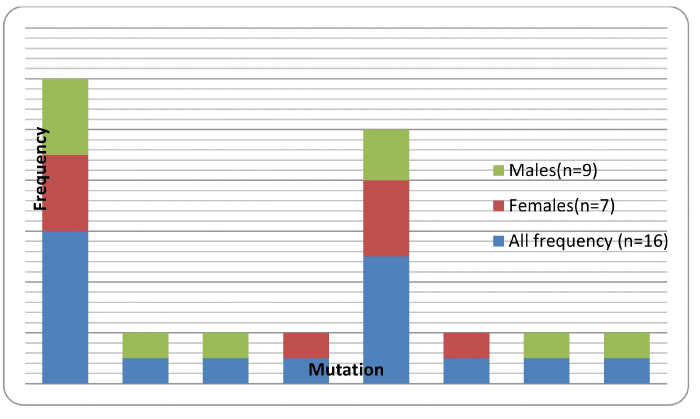
| Overall HIV associated drug resistant mutations prevalence in infant samples collected in the January 2010 to January 2011 period was 50% (16/32) and excluding the polymorphic mutations 25% (8/32) of the samples analyzed, The total number of infants recruited into the national EID program between January 2010 and January 2011 was 1 356. The prevalence of HIV infection reported in this study was 2.4% (32/1356) with most samples 97.6% (1324/1356) not meeting the study criteria (i.e HIV negative). Figure 1 summarizes the HIVDR mutation frequencies found in 16/32 infant samples analysed stratified by gender. One of the 16 HIVDR positive samples had at least two mutations K103KN and the Y181C. Of the sixteen patients with HIVDR, 5 had NVP only resistance, none had 3TC resistance and 8 had both etravirine (ETV) and rilpivirine (RPV) resistance, 2 had both NVP, efavirenz (EFV) and 1 had rilpivirine only (Table 1). The study shows an apparent NNRTI drug resistance. | |
| Discussion | |
| The current study reports a combined NNRTI drug resistance prevalence of 25%, a meta-analysis of 7 studies showed an NNRTI drug resistance range of 37-67% and an average of 52.6% [31]. In another study in the USA an NNRTI drug resistance prevalence of 23.8% among infants with a median age of 9.7 weeks was reported [30]. The current study reports a lower prevalence of NNRTI drug resistance compared to other African studies [38] and points to a rational use of NNRTIs in developed countries or the use of alternative protease inhibitor drugs. | |
| NVP drug resistance was 21.9 (7/32) in this study, in the metaanalysis by Arrive et al. the prevalence of NVP drug resistance was 35.7% [31]. This is similar to this study, and suggests a more careful approach to NVP use in resource poor settings that do not have access to alternative drugs. Worth noting is the absence of AZT drug resistance in this cohort, paradoxically the mothers had greater exposure to this drug compared to either NVP or 3TC ante-partum to post-partum. A study including a bigger sample of infants exposed to AZT ante-partum may best address this apparent lack of AZT resistance. Absence of resistance to previously used AZT does not rule out reservoirs of resistant virus that might emerge after re-initiation of the drug. This may suggest that the AZT resistant variants were less than 20% of the viral population and hence may not have been detected. | |
| E138A and Y181C mutations were the most common in our study group, 37.5% (6/16) and 31.3% (5/16) respectively. E138A is a polymorphic mutation that ranges in prevalence from 0.5% in subtype B viruses to 5% in subtype C viruses [39]. We report here a relatively high E138A mutation prevalence, which is weakly selected by ETR and RPV. The E138A mutation reduces ETR and RPV [39,40], susceptibility about 2-fold. Y181C is a non-polymorphic mutation selected in vitro by NVP, EFV, ETR and RPV [41-44]. The most clinically significant mutation is the Y181C mutation as it highly selected by NNRTIs. | |
| The study did not have information on whether the mothers had taken or been exposed to ETV or RPV because it is not mentioned in the database. It is reasonable to assume that during the option A era, ETV and RPV were relatively new drugs on the market and their distribution and use in low-income countries was not widely spread. The ETV and RPV resistance could have been as a result of another NNRTI use, NVP in this case. NVP over usage in that period were single dose NVP before labour was tailed by NVP daily for six weeks [29]. On the other hand the Y181C is a NVP resistance conferring mutation and its high prevalence is justifiable yet not acceptable during an option A era in a developing country. It will also be worthwhile doing a similar study using specimens from the current option B and B+ pMTCT programs as a way of HIV DR surveillance. | |
| The total number of infants recruited into the EID program between January 2010 and January 2011 was 1 356. The prevalence of HIV infection as reported in this study was 2.4%, this complies with the nations goal to reduce prevalence to less than 5%. In a study done in Cameroon on the EID cohort the prevalence of HIV infections among the 1423 infants included was 3.6%, comparable to this study [45]. A study on EID in Botswana reported a 7% HIV prevalence among 1931 infants recruited over 6months in 2005 [38], this high proportion may be justified because the EID and pMTCT programs were in their early days. HIV prevalence in most of the studies reviewed was <5% except for Botswana (7%), this may be an effect of the HIV prevalence in the adult population, 33.4% [38]. However, in a South African study to determine the acceptability and feasibility of universal HIV testing of infants attending immunization clinics to achieve early diagnosis of HIV and referral for HIV treatment and care services they reported an HIV prevalence of 9.2% [46]. | |
| Despite the revised 2010 WHO recommendations and the EID program many infants still get vertically transmitted HIV. Implementation of the EID program is not exactly complete in many developing countries [47]. In the Zimbabwean setting the complete EID process was complete for 80% (1356/ 1695) between January 2009 to January 2010. This was also true for Botswana were the EID process was complete for 81% (753/930) of the infants [38]. In Cameroon 83.9% of the initial participants completed the EID process [48], and in Tanzania it was 53%, 242 out of 441 initial participants enrolled [48] and in South Africa 65% of the participants completed the process [46]. | |
| Conclusion | |
| The study shows an apparent NNRTI drug resistance predominance and suggests a judicious use of NNRTI regimens and a prudent strategy to minimize the selection of drug resistance mutations. The authors recommend HIVDR surveillance in pregnant mothers and infants in resource limited settings. | |
| Acknowledgements | |
| The authors would like to acknowledge the help rendered by Justen Manasa, in providing technical expertise in the use and interpretation of the results of the sequencing system used. Special thanks also go to Professor David Katzenstein for his mentorship. This work was supported by Grant number 2U2RTW007367 from the Fogarty International Centre, National Institutes of Health (NIH, USA) through the International Clinical, Operational and Health Services and Training Award (ICOHRTA). | |
| Conflict of Interest | |
| The authors state no conflict of interest. | |
References
- Ledergerber B, Battegay M (2014) Epidemiology of HIV. TherUmsch: 2014 Aug; 2071(2018):2437-2041.
- WHO (2010) Antiretroviral drugs for treating pregnant women and preventing HIV infections in infants. Geneva, Switzerland: World Health Organization. p. 1-106.
- Shafer RW (2009) The challenge of antiretroviral drug resistance in HIV-1-infected children. J Pediatr (Rio J): 2009 Mar-Apr;2085(2002):2091-2004.
- Tudor Car L, van-Velthoven MH, Brusamento S, Elmoniry H, Car J, et al. (2011) Integrating prevention of mother-to-child HIV transmission (PMTCT) programmes with other health services for preventing HIV infection and improving HIV outcomes in developing countries. Cochrane Database Syst Rev: 2011 Jun 2015;(2016):CD008741.
- Bertagnolio S, Penazzato M, Jordan MR, Persaud D, Mofenson LM, et al. (2012) World Health Organization generic protocol to assess drug-resistant HIV among children <18 months of age and newly diagnosed with HIV in resource-limited countries. Clin Infect Dis 54 Suppl 4: S254-260.
- Mellors JW, Chow JY (2009) Single-dose nevirapine to prevent mother-to-child transmission of HIV type 1: balancing the benefits and risks. Clin Infect Dis 48: 473-475.
- Aurpibul L, Puthanakit T (2014) Review of Tenofovir Use in HIV-Infected Children. Pediatr Infect Dis J .
- Chokephaibulkit K, Cressey TR, Capparelli E, Sirisanthana V, Muresan P, et al. (2011) Pharmacokinetics and safety of a new paediatric fixed-dose combination of zidovudine/lamivudine/nevirapine in HIV-infected children. AntivirTher 16: 1287-1295.
- Phelps BR, Rakhmanina N (2011) Antiretroviral drugs in pediatric HIV-infected patients: pharmacokinetic and practical challenges. Paediatr Drugs 13: 175-192.
- Buchholz B, Hien S, Weichert S, Tenenbaum T (2010) Pediatric aspects of HIV1-infection--an overview. Minerva Pediatr 62: 371-387.
- Ubesie AC (2012) Pediatric HIV/AIDS in sub-Saharan Africa: emerging issues and way forward. Afr Health Sci 12: 297-304.
- Ahmed S, Kim MH, Sugandhi N, Phelps BR, Sabelli R, et al. (2013) Beyond early infant diagnosis: case finding strategies for identification of HIV-infected infants and children. AIDS 27 Suppl 2: S235-245.
- Adebimpe WO (2013) Challenges facing early infant diagnosis of HIV among infants in resource poor settings. Afr J Reprod Health 17: 122-129.
- Pilotto JH, Velasque LS, Friedman RK, Moreira RI, Veloso VG, et al. (2011) Maternal outcomes after HAART for the prevention of mother-to-child transmission in HIV-infected women in Brazil. AntivirTher 16: 349-356.
- Barral MF, de Oliveira GR, Lobato RC, Mendoza-Sassi RA, Martínez AM, et al. (2014) Risk factors of HIV-1 vertical transmission (VT) and the influence of antiretroviral therapy (ART) in pregnancy outcome. Rev Inst Med Trop Sao Paulo 56: 133-138.
- Volmink J, Siegfried NL, van der Merwe L, Brocklehurst P (2007) Antiretrovirals for reducing the risk of mother-to-child transmission of HIV infection. Cochrane Database Syst Rev : CD003510.
- Ciaranello AL, Perez F, Maruva M, Chu J, Engelsmann B, et al. (2011) WHO 2010 guidelines for prevention of mother-to-child HIV transmission in Zimbabwe: modeling clinical outcomes in infants and mothers. PLoS One 6: e20224.
- Wiegert K, Dinh TH, Mushavi A, Mugurungi O, Kilmarx PH (2014) Integration of prevention of mother-to-child transmission of HIV (PMTCT) postpartum services with other HIV care and treatment services within the maternal and child health setting in Zimbabwe, 2012. PLoS One 2014 Jun 10;9(6):e98236. doi: 10.1371/journal.pone.0098236.
- Sturt AS, Dokubo EK, Sint TT (2010) Antiretroviral therapy (ART) for treating HIV infection in ART-eligible pregnant women. Cochrane Database Syst Rev : CD008440.
- Ishikawa N, Shimbo T, Miyano S, Sikazwe I, Mwango A, et al. (2014) Health outcomes and cost impact of the new WHO 2013 guidelines on prevention of mother-to-child transmission of HIV in Zambia. PLoS One 9: e90991.
- Kuonza LR, Tshuma CD, Shambira GN, Tshimanga M (2010) Non-adherence to the single dose nevirapine regimen for the prevention of mother-to-child transmission of HIV in Bindura town, Zimbabwe: a cross-sectional analytic study. BMC Public Health 10: 218.
- Wagner TA, Frenkel LM (2010) Antiretroviral treatment for HIV-infected infants exposed to nevirapine. JAMA 304: 2589.
- Shafer RW (2006) Rationale and uses of a public HIV drug-resistance database. J Infect Dis 194 Suppl 1: S51-58.
- Giaquinto C Rampon O, De Rossi A (2006) Antiretroviral therapy for prevention of mother-to-child HIV transmission : focus on single-dose nevirapine. Clin Drug Investig 26: 611-627.
- Chersich MF, Gray GE (2005) Progress and Emerging Challenges in Preventing Mother-to-Child Transmission. Curr Infect Dis Rep 7: 393-400.
- Vairo F, Nicastri E, Liuzzi G, Chaula Z, Nguhuni B, et al. (2013) HIV-1 drug resistance in recently HIV-infected pregnant mother's naïve to antiretroviral therapy in Dodoma urban, Tanzania. BMC Infect Dis 13: 439.
- Lidström J, Li Q, Hoover DR, Kafulafula G, Mofenson LM, et al. (2010) Addition of extended zidovudine to extended nevirapine prophylaxis reduces nevirapine resistance in infants who were HIV-infected in utero. AIDS 24: 381-386.
- Moorthy A, Gupta A, Bhosale R, Tripathy S, Sastry J, et al. (2009) Nevirapine resistance and breast-milk HIV transmission: effects of single and extended-dose nevirapine prophylaxis in subtype C HIV-infected infants. PLoS One 4: e4096.
- Church JD, Omer SB, Guay LA, Huang W, Lidstrom J, et al. (2008) Analysis of nevirapine (NVP) resistance in Ugandan infants who were HIV infected despite receiving single-Dose (SD) NVP versus SD NVP plus daily NVP up to 6 weeks of age to prevent HIV vertical transmission. J Infect Dis 198: 1075-1082.
- Persaud D, Palumbo P, Ziemniak C, Chen J, Ray SC, et al. (2007) Early archiving and predominance of nonnucleoside reverse transcriptase inhibitor-resistant HIV-1 among recently infected infants born in the United States. J Infect Dis 195: 1402-1410.
- Arrivé E, Newell ML, Ekouevi DK, Chaix ML, Thiebaut R, et al. (2007) Prevalence of resistance to nevirapine in mothers and children after single-dose exposure to prevent vertical transmission of HIV-1: a meta-analysis. Int J Epidemiol 36: 1009-1021.
- Beck IA, Crowell C, Kittoe R, Bredell H, Machaba M, et al. (2008) Optimization of the oligonucleotide ligation assay, a rapid and inexpensive test for detection of HIV-1 drug resistance mutations, for non-North American variants. J Acquir Immune DeficSyndr 48: 418-427.
- Manasa J, Danaviah S Pillay S Padayachee P Mthiyane H et al. (2014) An affordable HIV-1 drug resistance monitoring method for resource limited settings. J Vis Exp .
- Drummond AJ, Suchard MA (2010) Bayesian random local clocks, or one rate to rule them all. BMC Biol 8: 114.
- Alcantara LC, Cassol S, Libin P, Deforche K, Pybus OG, et al. (2009) A standardized framework for accurate, high-throughput genotyping of recombinant and non-recombinant viral sequences. Nucleic Acids Res 37: W634-642.
- Gifford RJ, Liu TF, Rhee SY, Kiuchi M, Hue S, et al. (2009) The calibrated population resistance tool: standardized genotypic estimation of transmitted HIV-1 drug resistance. Bioinformatics 25: 1197-1198.
- de Oliveira T, Deforche K, Cassol S, Salminen M, Paraskevis D, et al. (2005) An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics 21: 3797-3800.
- Creek T, Tanuri A, Smith M, Seipone K, Smit M, et al. (2008) Early diagnosis of human immunodeficiency virus in infants using polymerase chain reaction on dried blood spots in Botswana's national program for prevention of mother-to-child transmission. Pediatr Infect Dis J 27: 22-26.
- Singh K, Marchand B, Rai DK, Sharma B, Michailidis E, et al. (2012) Biochemical mechanism of HIV-1 resistance to rilpivirine. J BiolChem 287: 38110-38123.
- Tambuyzer L, Nijs S, Daems B, Picchio G, Vingerhoets J (2011) Effect of mutations at position E138 in HIV-1 reverse transcriptase on phenotypic susceptibility and virologic response to etravirine. J Acquir Immune DeficSyndr 58: 18-22.
- Wu H Zhang HJ, Zhang XM, Xu HF, Wang M, et al. (2012) Identification of drug resistant mutations in HIV-1 CRF07_BC variants selected by nevirapine in vitro. PLoS One 7: e44333.
- Azijn H, Tirry I, Vingerhoets J, de Bethune MP, Kraus G, et al. (2010) TMC278, a next-generation nonnucleoside reverse transcriptase inhibitor (NNRTI), active against wild-type and NNRTI-resistant HIV-1. Antimicrob Agents Chemother 201054:718-727.
- Vingerhoets J Azijn H, Fransen E, De Baere I, Smeulders L, et al. (2005) TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J Virol 79: 12773-12782.
- Winslow DL Garber S, Reid C, Scarnati H, Baker D, et al. (1996) Selection conditions affect the evolution of specific mutations in the reverse transcriptase gene associated with resistance to DMP 266. AIDS 10: 1205-1209.
- Tejiokem MC Faye A, Penda IC, Guemkam G, AtebaNdongo F, et al. (2011) Feasibility of early infant diagnosis of HIV in resource-limited settings: the ANRS 12140-PEDIACAM study in Cameroon. PLoS One 6: e21840.
- Rollins N Mzolo S, Moodley T, Esterhuizen T, van Rooyen H (2009) Universal HIV testing of infants at immunization clinics: an acceptable and feasible approach for early infant diagnosis in high HIV prevalence settings. AIDS 23: 1851-1857.
- Cherutich P Inwani I, Nduati R, Mbori-Ngacha D (2008) Optimizing paediatric HIV care in Kenya: challenges in early infant diagnosis. Bull World Health Organ 86: 155-160.
- Nuwagaba-Biribonwoha H Werq-Semo B, Abdallah A, Cunningham A, Gamaliel JG, et al. (2010) Introducing a multi-site program for early diagnosis of HIV infection among HIV-exposed infants in Tanzania. BMC Pediatr 10: 44.
Tables and Figures at a glance
| Table 1 |
Figures at a glance
 |
| Figure 1 |
Relevant Topics
- Advanced Therapies
- Chicken Pox
- Ciprofloxacin
- Colon Infection
- Conjunctivitis
- Herpes Virus
- HIV and AIDS Research
- Human Papilloma Virus
- Infection
- Infection in Blood
- Infections Prevention
- Infectious Diseases in Children
- Influenza
- Liver Diseases
- Respiratory Tract Infections
- T Cell Lymphomatic Virus
- Treatment for Infectious Diseases
- Viral Encephalitis
- Yeast Infection
Recommended Journals
Article Tools
Article Usage
- Total views: 14832
- [From(publication date):
specialissue-2015 - Apr 02, 2025] - Breakdown by view type
- HTML page views : 10247
- PDF downloads : 4585
