Systematical Analysis to Assist the Significance of Rv1907c Gene with the Pathogenic Potentials of Mycobacterium tuberculosis H37Rv
Received: 03-Dec-2018 / Accepted Date: 27-Dec-2018 / Published Date: 05-Jan-2019 DOI: 10.4172/2155-952X.1000287
Abstract
Tuberculosis (TB) is currently among the major ten causes for death worldwide being brought about 1.7 million passing in 2016. The singular vaccine candidate available for its treatment till now is Bacillus Calmette-Guerin (BCG) which now also fails to provide accurate protection against this deadly disease. Mycobacterium tuberculosis H37Rv (M. tuberculosis H37Rv), the causative agent of this malady is currently turned out to be so intense and make extremely difficult to treat its infection. This manuscript covers some bioinformatics approaches against conserved hypothetical proteins which pretend to act as a thioredoxin-like protein. Thioredoxin (Trx) proteins are the redox proteins that are rotate between two forms, one is oxidized disulfide and the other one is reduced dithiol forms. Therefore it is said to be as an important component in many cellular redox reactions. The Trx protein of our study contains a conserved motif CXXC which is essential for its enzymatic activity. Mutational analysis of this protein declares its essentiality instability of the protein. This study concludes that this protein might be essential in several cellular processes carried out by this bacterium and further targeting this protein might give us a novel therapeutic antituberculosis drug.
Keywords: Mycobacterium tuberculosis H37Rv; Alveolar macrophages; Phagolysosome; Thioredoxin; Cysteine
Abbreviations
TB: Tuberculosis; M. tuberculosis H37Rv: Mycobacterium tuberculosis H37Rv; Trx: Thioredoxin; TrxR: Thioredoxin Reductase; GrxA: Glutaredoxin; FAD: Flavin Adenine Dinucleotide; NCBI: National Center for Biotechnology Information; MSA: Multiple Sequence Alignment; NADPH: Nicotinamide Adenine Dinucleotide Phosphate.
Introduction
Tuberculosis (TB) broadens its horizon day by day by increasing the numbers of individuals have been suffering from this disease [1]. Primarily this disease is caused by Mycobacterium tuberculosis H37Rv (M. tuberculosis H37Rv) in humans but there are other species also which contribute equally to infection in case of animals [2]. The pathogenesis due to this bacterium still resides as an escalating worldwide vigor problem that requires novel remedial and defensive procedures [3]. There are an estimated 10.4 million incidents of TB globally ranging from 8.8 million to 12.2 million were reported in 2016. According to World Health Organization (WHO) report TB burden and mortality rate is highest in India in comparison with other countries [4]. The facade of multi-drug-resistant strains of this bacterium is also creating a worldwide crisis. The confrontation of the bacterium in the cellular immune response plus their capability to endure as well as reactivation on an afterwards phase is scantily unstated [5].
M. tuberculosis H37Rv is an obligate aerobe which exists in the innate immune component of lung known as alveolar phagocytes which are mononuclear cells responsible for killing of most the pathogen but unfortunately is unable to affect M. tuberculosis H37Rv. These cells have been considered to employ a range of redox system to defend themselves adjacent to the oxidative stress produced through macrophages [6]. The above-stated studies have also exposed so as the thioredoxin scheme that might too assist for the upgrading of inactivated proteins originate through the oxidative stress come in front. Thioredoxin (Trx) proteins are the redox proteins that rotate between two forms, one is oxidized disulfide and the other one is reduced dithiol forms. Trx is in its oxidized state, in turn, showing reduction through flavoenzyme, thioredoxin reductase, through a redox-active cysteine conjugate plus flavin adenine dinucleotide like a substitute overwhelming cellular nicotinamide adenine dinucleotide phosphate (NADPH). Reduced Trx produced in this behavior is hence delivered for reducing disulfide of the intentioned proteins within the cellular environment [7]. The NADPH-dependent disulfide reduction method is necessarily used for a range of physiological roles.
For instance, two thioredoxin proteins are belonging to E. coli, TrxA and TrxC and another protein glutaredoxin have been recognized as the requirement for cellular DNA production through performing as an electron donor for the important enzyme ribonucleotide reductase. The thioredoxin system is consists of three components Trx, TrxR and NADPH. This system is an omnipresent redox conduit originates within the spacious assortment of phyla. The reduction of Trx that while present in their dithiol structure be the major disulfide reductase in living cells [8]. This redox scheme has an extensive assortment of the natural purpose, with sustaining the intracellular reduced condition in the facade of the oxidizing extracellular surroundings. Trx and TrxR inside M. tuberculosis H37Rv are probably participating comparable mean role opposing oxidative anxiety seeing already pragmatic in additional prokaryotes, consequently creating thioredoxin an objective intended for the antituberculosis drugs [9].
M. tuberculosis H37Rv whole genome sequence revealed that it encodes three thioredoxin proteins (TrxA, TrxR and TrxC). A recent report has been found the signaling of foremost is the biochemical inhibitor of M. tuberculosis H37Rv TrxC [10]. Since mycobacteria complex lacks the glutathione-dependent detoxification system, the thioredoxin scheme should importantly add for the pathogen resistance adjacent to oxidative intermediates and it is also shown that proteins are extremely preserved between living organisms like prokaryotes and eukaryotes, with sequence homology between 27 and 69% [11]. Correspondingly, M. tuberculosis H37Rv TrxC has 50 and 29% sequence homology to E. coli and human thioredoxin.
Trx system has been exposed to detoxify injurious agents, for example, H2O2, alkyl hydroperoxidase and supplementary oxidants that are also involved in preclusion, interference and the restore protein structure that somehow get damaged by oxidative stress [12]. The earlier abandoned reduction of glutathione disulfide by the thioredoxin system is currently being considered in the group; GSSG which may be able to attain considerable concentrations. In this circumstance of defense from oxidative tension, it is remarkable that the thioredoxin system might be an in-vitro electron donor for the so-called plasma glutathione peroxidase [13].
Reports on M. tuberculosis H37Rv relate the thioredoxin redox cycle with resistance to the isoniazid, the first line drug in TB treatment [14]. The function of the thioredoxin arrangement has also been examined in other protozoan parasites such as Entamoeba histolytica (E. histolytica). It is a facultative anaerobe an d can assault host tissues; where it is faced with the human immune response and required to deal with immediate oxygen species. A parasitic superoxide dismutase detoxifies superoxide anions unrestricted from activated macrophages. Hydrogen peroxide produced in this effect is consequently reduced by the mutual action of a thioredoxin reductase-like protein and a thioredoxin peroxidase-like protein. Recently it has been shown that these antioxidant enzymes are also dependable for the improvement of confrontation to metronidazole [15], a drug used for more than 25 years in the management of anaerobic and microaerophilic pathogens. However, the precise participation of the thioredoxin reductase-like enzyme in this method remains to be elucidated. Therefore in this manuscript, we show various bioinformatics aspects as structural, functional and mutational studies of gene Rv1907c of M. tuberculosis H37Rv which might empower experimental work in this field and might be found a suitable antituberculosis drug as shown in Table 1 [16-18].
| S.No. | Tools | URL | Function | Reference |
|---|---|---|---|---|
| 1 | Clustal Omega | https://www.ebi.ac.uk/Tools/msa/clustalo/ | For the Multiple sequence alignment. | [20] |
| 2 | STRING | https://string-db.org/cgi/network.pl?taskId=BUe3enVFzh8M | For the study of protein-protein interaction. | [21] |
| 3 | SignalP 4.1 | http://www.cbs.dtu.dk/services/SignalP/ | For the predicts the presence and location of signal peptide cleavage sites | [22,23] |
| 4 | DiANNA 1.1 | http://clavius.bc.edu/~clotelab/DiANNA/ | For the study of Cysteine state and Disulfide bond partner prediction. | [25,26] |
| 5 | PSIPRED v3.3 | http://bioinf.cs.ucl.ac.uk/psipred/ | For predict secondary structure of the protein. | [27,28] |
| 6 | iSTABLE | http://predictor.nchu.edu.tw/istable/ | For the integrated predictor for protein stability change upon single mutation. | [30] |
| 7 | DynaMut webserver | http://biosig.unimelb.edu.au/dynamut/analysis | For the prediction of protein stability changes upon mutation using Normal Mode Analysis | [31] |
Table 1: Table showing enlists the different servers used in the study.
Materials and Methods
Retrieval of the target protein sequence
Mycobrowser is the database where M. tuberculosis H37Rv data is available easily like nucleotide sequence or protein sequence else physiochemical properties are also easily available here. For the protein sequence, we know the enzymatic functions, signaling functions, structural functions etc. We have selected the sequence of the Rv1907c hypothetical gene for the study of the thioredoxin-like protein [19].
Multiple sequence alignment
MSA is the alignment of three or more biological sequences of similar length. The applications of the multiple sequence alignment outcome, evolutionary relationship between the sequences, homology can be evidenced studies. For generating the sequence alignment Clustal Omega uses HMM profile-profile techniques [20]. Clustal Omega tool is suitable for medium-large alignments. Biological sequence similarity by Pairwise Sequence Alignment tool is an important step for identifying the relationship of functional, structural and/or evolutionary characteristics.
Protein-protein interaction network
For the study of the protein-protein interaction between two genes, there will be a server known as STRING database. Protein-protein interaction connections as direct (physical) or indirect (functional) associations; String could do that from computational prediction; from in sequence communication between organisms, and from connections aggregated from other principal databases [21]. String database interaction result shown separately for each data and the cutoff value as low confidence: scores <0.4; medium: 0.4 to 0.7; high: >0.7.
Prediction of the signal peptide
Analysis of cleavage site prediction was done by SignalP 4.1 server. Rv1907c protein sequence was examined by SignalP 4.1 server (http://www.cbs.dtu.dk/-services/SignalP). The Y-score (combined cleavage site score) distinguishes between C-score peaks by choosing the one where the slope of the S-score is steep. The S-score (signal peptide score) distinguish positions within signal peptides from positions in the mature part of the proteins and from proteins without signal peptides and the C-score is trained to be high at the position immediately after the cleavage site [22-24].
Disulfide-bonding in protein
Disulfide bonds in protein play an important role in protein stability conformation. Primary structure of protein cysteine residue formed disulfide bridges. Disulfide bonds and Cysteine residues are important for protein structure and function. Due to the utilization of SVM binary server to DiANNA 1.1 web server predict bonding state of cysteine, these cysteines are paired by Recursive Neural Network to show disulfide bridges [25,26]. Study of the disulphide bonding helps for experimental structure determination and defining the stability of the proteins.
Secondary structure prediction
The secondary structure was predicted by using PSIPRED v3.3 (http://bioinf.cs.ucl.ac.uk) servers [27,28].
Mutational analysis
In proteomics and genomics, contemplates studies of protein strength free energy change (ΔΔG) upon single point mutation may likewise enable for the explanation to process. The greater part of the ΔΔG esteems are almost zero (around 32% of the ΔΔG informational index ranges from −0.5 to 0.5 kcal/mole) and both the esteem and indication of ΔΔG might be either positive or negative for a similar change obscuring the relationship among mutated and expected ΔΔG esteem. Keeping in mind the final goal to conquer this issue, we portray another indicator that segregates between 3 change classes: destabilizing mutations (ΔΔG<−1.0 kcal/mol), balancing out transformations (ΔΔG>1.0 kcal/mole) and nonpartisan changes (−1.0 ≤ ΔΔG ≤ 1.0 kcal/ mole) [29]. Prediction of protein stability change upon a single point mutation was predicted by iStable online server [30] and DynaMut web server [31].
Results
Retrieval of the target protein sequence
For the retrieval of the target protein sequence, we have been used Mycobrowser database which has all the nucleotide and protein sequence of M. tuberculosis H37Rv strain. Our query protein Rv1907c has 648 bp gene length or 24 kDa protein. It is a conserved hypothetical protein and not studied before.
A Multiple sequence alignment
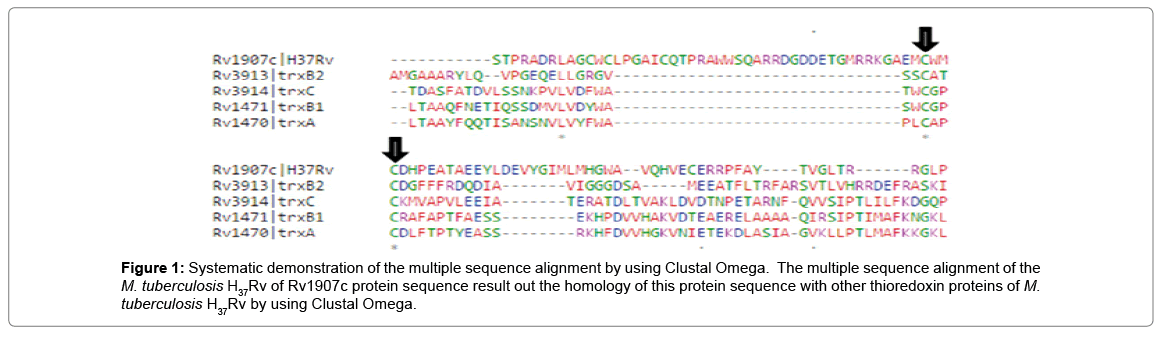
In multiple sequence alignment tool Clustal Omega, we studied our query protein Rv1907c of M. tuberculosis H37Rv shows the homology with other conserved thioredoxin family protein of M. tuberculosis H37Rv as shown in Figure 1.
Figure 1: Systematic demonstration of the multiple sequence alignment by using Clustal Omega. The multiple sequence alignment of the M. tuberculosis H37Rv of Rv1907c protein sequence result out the homology of this protein sequence with other thioredoxin proteins of M. tuberculosis H37Rv by using Clustal Omega.
Protein-protein interaction network
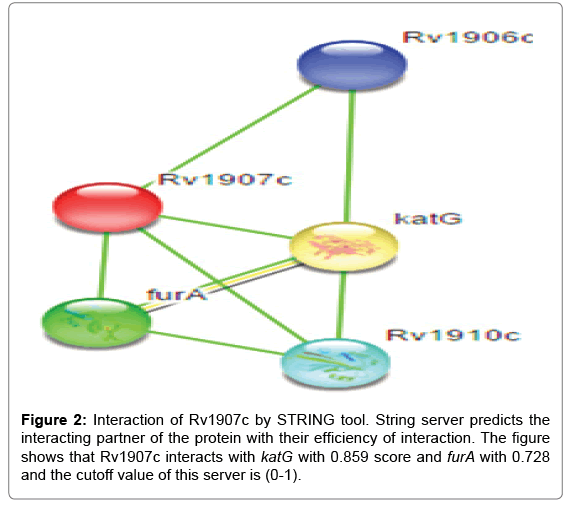
Inside the environment of cell type protein-protein interaction with other proteins, In silico tools STRING v9.1 have been used for this study. STRING is a search tool for retrieval of interacting genes. A study of a large repository of protein-protein interactions has been done by STRING database; involving functional interactions, stable complexes, and regulatory interactions along with proteins. The cutoff value of STRING server is between (0-1), low confidence: scores <0.4; medium: 0.4 to 0.7; high: >0.7. The results of Rv1907c protein-protein interaction shows that it is highly interacted with the katG and furA genes. These both genes present in immediate upstream region of the Rv1907c and thus highly interact with interested protein. katG gene is involved in non protective oxidative stress response and furA is found to be negative regulator of katG gene. For the functioning partner katG act as a bi-functional enzyme with both catalase and broad-spectrum peroxidase activity and furA shows ferric uptake regulation protein as shown in Figure 2, better consideration of the interaction networks should be seen on the server site.
Prediction of the signal peptide
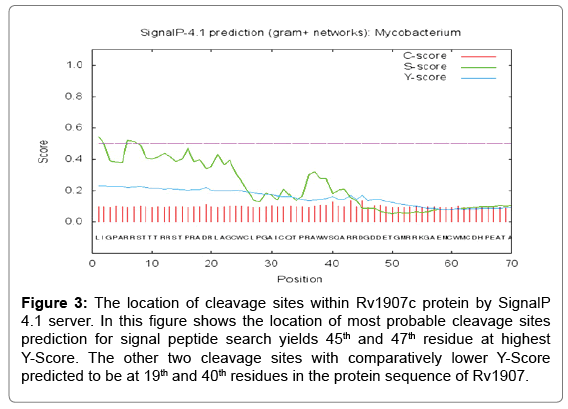
Signal peptide prediction was done by SignalP 4.1 prediction server. In the SignalP 4.1 server, FASTA format of the protein sequence is acceptable and other criteria are fulfill for running the program are organism group, D-cutoff value, graphical output, output format, method and positional limit. In our study, the organism group is Gram-positive bacteria with the D-Score cut-off value of 0.45. For the searching of signal peptide result are 45th residue at highest Y-Score and therefore the other two cleavage sites with relatively lower Y-Score predicted to be as 45th and 47th position residue is in the protein sequence of Rv1907c. All the cleavage sites were predicted towards the carboxyl- terminal of arginine as shown in Figure 3.
Figure 3: The location of cleavage sites within Rv1907c protein by SignalP 4.1 server. In this figure shows the location of most probable cleavage sites prediction for signal peptide search yields 45th and 47th residue at highest Y-Score. The other two cleavage sites with comparatively lower Y-Score predicted to be at 19th and 40th residues in the protein sequence of Rv1907.
Disulfide-bonding in protein
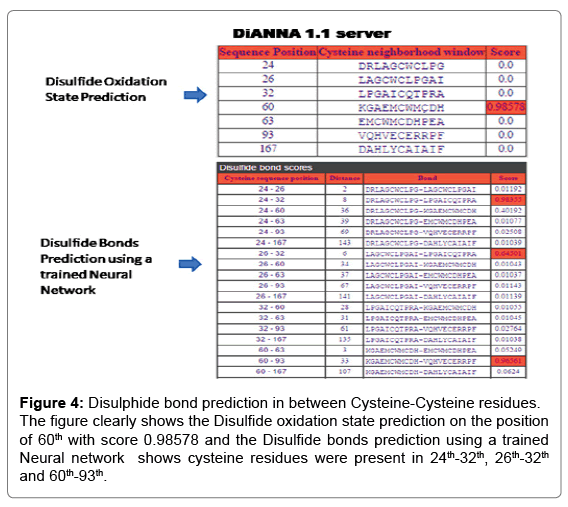
For the analysis of Rv1907c protein by DiANNA 1.1 web server as shown in Figure 4 shows the Disulfide oxidation state prediction on the 60th position with score 0.98578 and the Disulfide bond prediction on the position of 24th-32th, 26th-32th and 60th-93th.
Figure 4: Disulphide bond prediction in between Cysteine-Cysteine residues. The figure clearly shows the Disulfide oxidation state prediction on the position of 60th with score 0.98578 and the Disulfide bonds prediction using a trained Neural network shows cysteine residues were present in 24th-32th, 26th-32th and 60th-93th.
Secondary structure prediction
PSIPRED server predicted to the secondary structure prediction of the protein analysis by the single amino acid sequence or FASTA format submission of the protein sequence (https://www.predictprotein.org/). For the analysis of PSIPRED server predicted the Helix (H), extended strand in beta-sheet (E) and coiled region of the protein. The viewer layout predicted features that correspond to regions of the Rv1907c protein queried sequence. The positions of the amino acids show their protein binding region as well as polynucleotide binding regions respectively as shown in Figure 5. Whereas the PSIPRED server has predicted that the events of the 4 α-helices and 6 β-strands are shown in the Rv1907c protein sequence.
Mutational analysis
iStable metaserver and STRUM are utilized for the analysis of mutational studies of our query protein to analyze the single point mutation. In Rv1907c which is hypothetical protein has a thioredoxin motif (CXXC). For the single point mutation studies, we have selected cysteine residue because of the CXXC motif and cysteine residue make disulfide bond which is important for experimental structure determination and defining the stability of the proteins. In Rv1907c, there are seven cysteine residues present on 24, 26, 32, 60, 63, 93 and 167 positions. Moreover, cysteine residue 60th and 63rd position are the thioredoxin motif site, therefore, it was decided to check change in stability of the protein at this cysteine.
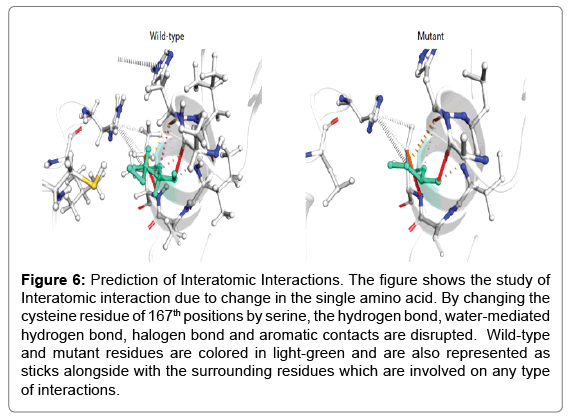
Single point mutation studies through protein sequence: In thioredoxin protein Cysteine residue is crucial for functional activity, therefore mutation at this position with all other amino acid had been shown in Figure 6. iStable meta server calculation there are DDG<- 0.5 means (large decrease of stability), DDG>0.5 means (increase of stability) and -0.5<=DDG<=0.5 means (neutral stability). In the graph, we have seen that there is more than 3 amino acids have largely decreased the stability means -0.5 or below value towards negative (Tables 2 and 3). In the past research analysis, there were some findings which prove the mutation of cysteine by serine shows the major function loss. As further studies in thioredoxin, protein motif is also CXXC so we select the cysteine residue for mutation. In the iStable metaserver program we select a 25°C temperature and 7.0 pH.
Figure 6: Prediction of Interatomic Interactions. The figure shows the study of Interatomic interaction due to change in the single amino acid. By changing the cysteine residue of 167th positions by serine, the hydrogen bond, water-mediated hydrogen bond, halogen bond and aromatic contacts are disrupted. Wild-type and mutant residues are colored in light-green and are also represented as sticks alongside with the surrounding residues which are involved on any type of interactions.
| Position | I-Mutant 2.0 | MUpro | iStable | |||
|---|---|---|---|---|---|---|
| DDG | Stability | DDG | Conf. Score | Stability | Conf. Score | |
| C24S | -0.58 | Decrease | -1.28 | -0.99006162 | Decrease | 0.774014 |
| C26S | -0.58 | Decrease | -1.04 | -0.78773185 | Decrease | 0.783164 |
| C32S | -0.33 | Decrease | -1.89 | -0.67104070 | Decrease | 0.795649 |
| C60S | -0.63 | Decrease | -1.13 | -0.66134712 | Decrease | 0.816418 |
| C63S | -0.53 | Decrease | -1.58 | -1 | Decrease | 0.820475 |
| C93S | -0.71 | Decrease | -1.01 | -0.93314897 | Decrease | 0.671629 |
| C167S | -0.73 | Decrease | -1.28 | -0.37210931 | Decrease | 0.816823 |
Table 2: Comparative studies by iStable (Metaserver) for protein stability prediction confirms that the wild type cysteine on position 167th mutate with serine (Cys167Ser) has highest decrease in stability.
| Position | DynaMut | ENCoM | mCSM | SDM | DUET |
| DDG value/ | DDG value/ | DDG value/ | DDG value/ | DDG value/ | |
| Stability | Stability | Stability | Stability | Stability | |
| C24S | -0.297 Decrease | -0.289 Decrease | -1.790 Decrease | -1.310 Decrease | -1.708 Decrease |
| C26S | 0.017 Increase | 0.227 Increase | -1.806 Decrease | -1.400 Decrease | -1.756 Decrease |
| C32S | -0.087 Decrease | -0.504 Decrease | -1.107 Decrease | -1.850 Decrease | -1.102 Decrease |
| C60S | -0.367 Decrease | -0.255 Decrease | -0.110 Decrease | -0.730 Decrease | -0.174 Decrease |
| C63S | -0.272 Decrease | -0.282 Decrease | -0.893 Decrease | -1.350 Decrease | -0.770 Decrease |
| C93S | -0.425 Decrease | -0.433 Decrease | -0.917 Decrease | -1.940 Decrease | -1.052 Decrease |
| C167S | -1.560 Decrease | -0.581 Decrease | -2.213 Decrease | -1.230 Decrease | -2.109 Decrease |
Table 3: Study of protein stability change after a single point mutation by using Normal Mode Analysis by DynaMut server. The table concludes that the stability highest decreases stability at 167th residue by changing cysteine to serine.
Protein stability changes upon mutation using normal mode analysis by DynaMut server: DynaMut web server has been used for the analysis of the impact of the mutations on protein dynamics and stability resulting from vibration entropy changes. This web server executes two distinct, normal mode approaches which can be used to analyze and visualize protein dynamics by sampling conformations and their assessment. DynaMut integrates our graph-based marks alongside through usual mode dynamics to produce a consensus prediction of the impact of the mutation on protein stability. This approach outperforms alternative approaches to predict the effects of mutations on protein stability and flexibility (p-value <0.001), achieving a correlation of up to 0.70 on blind tests. Proteins are extremely vibrant molecules, whose function is fundamentally linked to their molecular motions. In spite of the crucial role of protein dynamics, their computational simulation cost has led to most structure-based approaches for evaluating the impact of mutations on protein structure and function relying upon static structures.
Discussion
Studies regarding these findings lead us to know about the risk of development of this disease in new individuals. The risk is on rising due to unavailability of an effective drug. Although the number of incidences had been decreased, the risk increases day by day also due to several environmental factors. There is continuous effort had been put by scientists in order to increase the effectiveness of the vaccine and in searching for a new drug target. This manuscript elaborates the characteristics of the Rv1907c gene of M. tuberculosis H37Rv. This 24kDa protein is a conserved hypothetical gene [17]. Multiple sequence alignment data showed that this gene share similarity with other thioredoxin genes of M. tuberculosis H37Rv which might be an important finding towards its effectiveness as a thioredoxin protein [18]. STRING database shows its several interacting partners which include its neighboring partners as well as katG and furA [19]. These both genes are present in adjacent upstream region of Rv1907c and katG involved in response from non protective oxidative stress process occurring in M. tuberculosis . furA is found to be negatively regulating expression of katG gene. The study of these two genes thus remains us willing to knowing the fact about involvement of Rv1907c in stress response [32,33]. SignalP 4.1 server has been used for the prediction of cleavage site present in Rv1907c. This server shows the location of the most probable cleavage site at 45th and 47th residue with highest y score. There are two other cleavage sites which are lower than the Y-Score to be predicted as 19th and 40th residues in the protein sequence of Rv1907c [22-24]. Disulfides bonding are also present in 24th-32th, 26th-32th and 60th-93th position in the protein [25,26]. For the secondary structure prediction of Rv1907c protein sequence finds 4 alpha helices (from 36th-44th, 70th-83th, 118th-134th and 163rd-172th) and 6 beta sheets [27,28]. Thioredoxin proteins contain a conserved amino acid motif CXXC which is essential for its activity and cysteine residue make disulfide bond which is important for experimental structure determination and defining the stability of the proteins [29]. Cysteine residue 60th and 63rd position are the thioredoxin motif site, therefore, we check to change the stability of the protein at this cysteine. Single point mutation by the iStable metaserver proves that the mutation of cysteine at 167th position (C167S) with serine causes maximum loss of stability [30] and also we cross-check with DynaMut server finds a loss of interatomic interaction in the mutant type in comparison with the wild-type species [31]. After analysis of Rv1907c, we find that this gene might possess’ thioredoxin activity that seems to be essential for the vitality of M. tuberculosis H37Rv inside the phagosomes of the host. Further study of this gene might come out with the production of effective drug therapy against this deadly disease.
Conclusion
The Trx protein of our study contains a conserved motif CXXC which is essential for its enzymatic activity. Mutational analysis of this protein declares its essentiality instability of the protein. This study concludes that this protein might be essential in several cellular processes carried out by this bacterium and further targeting this protein might give us a novel therapeutic antituberculosis drug.
Acknowledgement
The authors acknowledge financial support from the Department of Science and Technology-SERB, Council of Scientific and Industrial Research-Institute of Genomics and Integrative Biology under the research project GAP0145.
References
- Meena LS, Rajni (2010) Survival mechanisms of pathogenic Mycobacterium tuberculosis H37Rv. FEBS J 277: 2416-2427.
- Shivangi, Meena LS (2018) A Novel Approach in Treatment of Tuberculosis by Targeting Drugs to Infected Macrophages Using Biodegradable Nanoparticles. Appl Biochem Biotechnol 185: 815-821.
- Glickman MS, Jacobs WR (2001) Microbial pathogenesis of Mycobacterium tuberculosis: dawn of a discipline. Cell 104: 477-485.
- Akif M, Suhre K, Verma C, Mande SC (2005) Conformational flexibility of Mycobacterium tuberculosis thioredoxin reductase: crystal structure and normal-mode analysis. Acta Crystallogr D Biol Crystallogr 61: 1603-1611.
- Shinnick TM, King CH, Quinn FD (1995) Molecular biology, virulence, and pathogenicity of mycobacteria. Am J Med Sci 309: 92-98.
- Holmgren A (2000) Antioxidant function of thioredoxin and glutaredoxin systems. Antioxid Redox Signal 2: 811-820.
- Gleason FK, Holmgren A (1988) Thioredoxin and related proteins in procaryotes. FEMS Microbiol Rev 4: 271-297.
- Gasdaska JR, Berggren M, Powis G (1995) Cell growth stimulation by the redox protein thioredoxin occurs by a novel helper mechanism. Cell Growth Differ 6: 1643-1650.
- Zhang Z, Hillas PJ, Ortiz PR (1999) Reduction of peroxides and dinitrobenzenes by Mycobacterium tuberculosis thioredoxin and thioredoxin reductase. Arch Biochem Biophys 363: 19-26.
- Hall G, Shah M, McEwan PA, Laughton C, Stevens M, et al. (2006) Structure of Mycobacterium tuberculosis thioredoxin C. Acta Crystallogr D Biol Crystallogr 62: 1453-1457.
- Eklund H, Gleason FK, Holmgren A (1991) Structural and functional relations among thioredoxins of different species. Proteins 11: 13-28.
- Gromer S, Schirmer RH, Becker K (1999) News and views on thioredoxin reductases. Redox Rep 4: 221-228.
- Bjornstedt M, Xue J, Huang W, Akesson B, Holmgren A (1994) The thioredoxin and glutaredoxin systems are efficient electron donors to human plasma glutathione peroxidase. J Biol Chem 269: 29382-29384.
- Volkman HE, Clay H, Beery D, Chang CW, Sherman DR, et al. (2004) Tuberculous Granuloma Formation Is Enhanced by a Mycobacterium Virulence Determinant. PLoS Biol 2: e367.
- Wassmann C, Hellberg A, Tannich E, Bruchhaus I (1999) Metronidazole resistance in the protozoan parasite Entamoeba histolytica is associated with increased expression of iron-containing superoxide dismutase and peroxiredoxin and decreased expression of ferredoxin and flavin reductase. J Biol Chem 274: 26051-26056.
- Beg MA, Shivangi, Thakur SC, Meena LS (2018) Structural prediction and mutational analysis of Rv3906c gene of Mycobacterium tuberculosis H37Rv to determine its essentiality in survival. Adv in Bioinfo 1: 1-12.
- Shivangi, Beg A, Meena S, Meena LS (2017) To Find out the Essentiality of Rv0526 Gene in Virulence of Mycobacterium tuberculosis by using In silico Approaches. Open J Bac 1: 013-015.
- Kapopoulou A, Lew JM, Cole ST (2011) The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculo 91: 8-13.
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, et al. (2011) Fast, saclable generation of high quality protein multiple sequence alignment using Clustal Omega. Mol Syst Biol 7: 539.
- Lewis AC, Saeed R, Deane CM (2010) Predicting protein-protein interactions in the context of protein evolution. Mol Biosyst 6: 55-64.
- Nielsen H, Engelbrecht J, Brunak S, von HG (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavages sites. Protein Eng Des Sel 10: 1-6.
- Bendtsen JD, Nielsen H, von HG, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783-795.
- Monu, Meena LS (2016) Biochemical characterization of PE_PGRS61 family protein of Mycobacterium tuberculosis H37Rv reveals the binding ability to fibronectin. Iran J Basic Med Sci 19: 1105-1113.
- Ferre F, Clote P (2005) DiANNA: A web server for disulfide connectivity prediction. Nucleic Acid Res 33: W230-W232.
- Ferre F, Clote P (2006) DiANNA 1.1: an extension of the DiANNA web server for ternary cysteine classification. Nucleic Acid Res 34: W182-W185.
- McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinfo 16: 404-405.
- Jones DT (2000) Protein structure prediction in the postgenomic era. Curr Opin Struct Biol 10: 371-379.
- Capriotti E, Fariselli P, Capriotti EE, Fariselli P, Calabrese R, et al. (2005) Predicting protein stability changes from sequences using support vector machines. Bioinfo 21: ii54-ii58.
- Chen CW, Lin J, Chu YW (2013) iStable: off-the-shelf predictor integration for predicting protein stability changes. BMC Bioinfo 14: S5.
- Rodrigues CH, Pires DE, Ascher DB (2018) DynaMut: predicting the impact of mutation on protein conformation, flexibility and stability. Nucleic Acid Res 46: W350-W355.
- Milano A, Forti F, Sala C, Riccardi G, Ghisotti D (2001) Transcriptional regulation of furA and katG upon oxidative stress in Mycobacterium smegmatis. J Bacteriol 183: 6801-6806.
- Zahrt TC, Song J, Siple J, Deretic V (2001) Mycobacterial FurA is a negative regulator of catalase-peroxidase gene katG. Mol Microbiol 39: 1174-1185.
Citation: Beg A, Shivangi, Thakur SC and Meena LS (2019) Systematical Analysis to Assist the Significance of Rv1907c Gene with the Pathogenic Potentials of Mycobacterium tuberculosis H37Rv. J Biotechnol Biomater 8:287. DOI: 10.4172/2155-952X.1000287
Copyright: © 2019 Beg A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 5231
- [From(publication date): 0-2018 - Nov 19, 2025]
- Breakdown by view type
- HTML page views: 4141
- PDF downloads: 1090