Systematic Analysis of GWAS Data Reveals Genomic Hotspots for Shared Mechanisms between Neurodegenerative Diseases
Received: 24-Jul-2017 / Accepted Date: 28-Nov-2017 / Published Date: 04-Sep-2017 DOI: 10.4172/2161-0460.1000368
Abstract
Objective: In this study, we have tried to reveal molecular mechanisms underlying “shared genetic variants” and developed a strategy to identify candidate mechanisms for shared aetiology of a pair of diseases, to uncover biological relationships between quantitative traits or related neurodegenerative diseases.
Methods: Genetic variants were collected from GWAS catalog, belonged to multiple disease association studies. Meta-analysis was performed by using Metal (a whole genome association analysis toolset), and normalized them for their different sample sizes. LD analysis was done with Haploreg DB V.4.0. Subsequently, the ENSEMBL variant database was used as a reference database. Additionally, these shared SNPs were interpreted with Regulome DB V.1.1 and finally ranked the variant lists according to predicted functional consequences attributes. Afterwards evidences were collected from gene expression studies, patents, knock-out studies and other literature.
Results: Pair-wise analysis also revealed that AD and PD have the largest number of shared disease-associated loci. Additionally, tau locus is discovered in a very novel and unique perspective of stress induced shared pathology of AD and PD, which provides suggestive evidence that the molecular mechanisms influencing aetiology and progression of selective neurodegenerative diseases are at least partly interrelated.
Conclusion: Genetic overlap between these diseases suggests that genomic locus should be considered to investigate the effects of GWAS variants rather than individual genetic variants, particularly to investigate shared pathology.
Keywords: GWAS; LD (Linkage Disequilibrium); Shared genetic loci; Genetic variants; Shared pathology; Neurodegenerative diseases
Background
Genome wide association studies have been very useful for the identification of genetic variants as disease risk markers; however, the impact of these genetic variants in disease aetiology remains largely unclear. In this study, we tried to unravel molecular mechanisms underlying “shared genetic variants” and developed a strategy to identify candidate mechanisms for shared aetiology of diseases that display similar patterns of genetic variation organized in shared genomic hotspots. We demonstrate how this approach leads to new insights that help to uncover biological relationships between quantitative traits or related neurodegenerative diseases.
Many traits or diseases have been shown to share genetic architecture [1,2]. This phenomenon, that a genetic variant affects multiple phenotypes, is often called ‘pleiotropy’ [3-5]. Such pleiotropic variants are particularly interesting, as the functional impact of a SNP on one or several genes may provide clues about the underlying molecular mechanism. For example, a significant overlap of shared genetic variants and pathways has been detected in immune-mediated diseases, suggesting extensive pleiotropic effects [6-8]. These shared genetics variants linked to pathways are ideally suited to identify candidate mechanisms underlying a “shared aetiology” of different diseases.
So far, various studies have been implemented across the genome, mostly on those groups of diseases, which are already well recognized or hypothesized to be interconnected [6,8-10] or by investigating influence of individual genetic variant on a wide range of diverse diseases [11-13].
Biologically, a genetic variant can influence different traits fundamentally in two different ways; firstly, it can influence two distinct phenotypes through two independent physiological mechanisms, while secondly, its effect on the second trait can be mediated through its effect on the first one.
Apparent genetic similarities in a pair of distinct diseases may be indicative for potential overlaps in the underlying disease mechanisms. Thus investigating common factors and network modules shared within a pair of distinct, but related diseases, may point at shared mechanisms. Rather than studying individual diseases separately, investigation and analysis of common dysregulated pathways or dysfunctional proteins of a pair of related diseases can be expected to reveal deeper comprehensive knowledge about pathophysiological processes.
Correspondingly, computing of shared molecular level mechanisms of related disorders can not only assist understanding of the etiology of a disease; but also such associations between shared pathways and correlation with biological processes can accelerate drug discovery efforts by suggesting promising treatment candidates for already approved drugs (known as drug repositioning) [14].
In the work presented here, we performed a systematic and comprehensive analysis of shared genomic loci likely to represent genomic hotspots with genes functionally involved in the aetiology of neurodegenerative diseases. We go way beyond classical meta-analysis of GWAS data by performing a ‘functional context enrichment’ that is tailored to embed candidate genes in these genomic hotspots in a mechanistic context. We demonstrate that this functional enrichment can lead to the identification of new candidate mechanisms for shared aetiology of Alzheimer´s Disease and Parkinsonism.
Methods
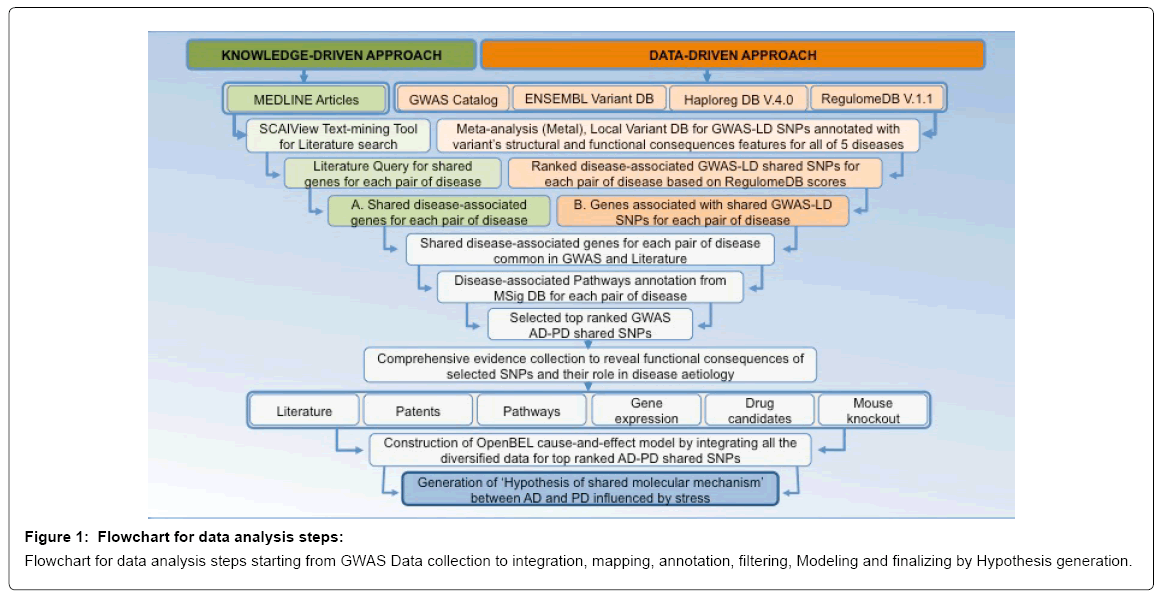
GWAS disease-associated variants are identified throughout the entire genome. In order to reveal shared genomic hotspots, that could have been comprised candidate genes for shared molecular mechanisms between two or multiple neurodegenerative and related diseases, genetic variants were collected from GWAS catalog [15] with the threshold of p-values-5 for five diseases; including Alzheimer’s disease, Parkinson disease, Schizophrenia, Multiple sclerosis and Type 2 diabetes mellitus. These collected genetic variants were belonged to multiple disease association studies and each association study was conducted with different sample sizes. Thus according to basic principle of meta-analysis, we combined the evidence for association from individual studies, with the implementation of appropriate weights, by using a whole genome association analysis toolset Metal [16] and normalized them for their different sample sizes.
Afterwards, Linkage Disequilibrium (LD) analysis was conducted separately for each disease by using Haploreg DB V.4.0 [17]. Next, shared genetic variants were queried by pair-wise analysis for ten pairs of disease of these five diseases.
Subsequently, we made use of the ENSEMBL variant database [18-20] as a reference database to map the SNPs with their relevant chromosome, location, gene, allele and potential functional features (intergenic SNPs were mapped to the nearest gene on the chromosome). Additionally, these shared SNPs were interpreted with the characteristics of predicted functional consequences by using RegulomeDB V.1.1 [21] to get annotation from current ENCODE data (updated with recent ENCODE releases: [22,23]), Chromatin States data from the Roadmap Epigenome Consortium and updated data for DNase footprinting, PWMs, and DNA Methylation, and finally ranked the variant lists according to predicted functional consequences attributes.
Most of the GWAS identified genetic variants are located on the non-coding regions of the genome. In order to investigate, whether there are any overlapping genome stretches between the ‘loci of shared GWAS and LD genetic variants’ and ‘loci of the well-established disease-associated genes in the literature’; in addition to the datadriven approaches described above, a comprehensive knowledge driven approach was also conducted, by searching systematically from literature with the help of a literature mining environment-SCAIView [24].
To extract shared genes for a pair of disease from literature, we were queried via SCAIView for those genes, which were studied for both diseases comprised in a pair (i.e., for AD and T2DM disease pair: {(([MeSH Disease:”Alzheimer Disease”]) and [MeSH Disease:”Diabetes Mellitus Type 2”]) and [Human Genes/Proteins]}). This literature search was conducted in a pair-wise analysis of genes for all of the ten pairs of diseases. The extracted list of “shared genes for a pair of disease from literature” (represented in the workflow as ‘List: A’) from SCAIView, was then used to pinpoint overlaps by comparing it with the list of “genes mapped with shared GWAS-LD genetic variants for a pair of disease” (represented in the workflow as ‘List: B’); and resulting file had ‘shared genes for a pair of disease’ common in GWAS-LD and Literature (Figure 1).
Afterwards, we mapped list of these shared genes to biological pathways by using MsigDB [25], to identify common pathways for each pair of disease. To demonstrate the potential of the approach, we did an exploratory study on one putative shared mechanism relevant for AD and PD. The genomics locus investigated maps to chromosome 17; to a region that displays highest scores for functional consequences in RegulomeDB and one of high ranked shared pathway between AD and PD from MsigDB result table, that is ‘KEGG_LONG_TERM_ DEPRESSION’ (Supplementary File). The high-resolution analysis of that shared genomic locus for its potential role in the aetiology of the disease pair AD and PD includes - besides the identification of the candidate locus and the candidate genes within - the collection of evidences from gene expression studies, patents, knock-out studies and other literature, ultimately resulting in a comprehensive knowledgedriven approach towards the enrichment with supportive evidence.
Results
In an initial step, we selected five different brain diseases including Alzheimer’s disease (AD), Parkinson disease (PD), Schizophrenia, Multiple sclerosis (MS) and Type 2 diabetes mellitus (T2DM).
Spatial analysis, after mapping of GWAS disease-associated intronic SNPs to the genes, they belong to; and intergenic SNPs to the most likely, nearby genes; reveals that most of the GWAS SNPs are located around specific genome loci (“genomic hotspots”). Our assumption is, that the genes existing in the vicinity of these genome loci may play a role in the dysregulation of disease-associated pathways.
Moreover, we computed pair-wise analysis for shared genetic variants to see the relevancy between each pair of diseases. However, enumerating of pair-wise shared GWAS SNPs before LD SNPs enrichment revealed that only a very limited number of individual SNPs are shared in a pair of diseases, while after LD analysis, most of the disease pairs showed a substantial count of shared variants; which also signify the genetically linkage between SNPs located on these specific genomic loci around GWAS SNPs. Thus it can be explained that these pairs of disease may share disease-associated genomic loci rather than individual genetic variants (Supplementary File).
Pair-wise analysis also revealed that AD and PD have the largest number of shared disease-associated loci. There is no doubt, that this is reflecting the bias that comes with the higher number of GWAS studies and available data around these two diseases. But it also may indicate an overlap of the genetics relevant for pathophysiology mechanisms shared between AD and PD. Other disease pairs, for instance the AD-T2DM pair, did also show a promising number of shared genetic markers and genomic loci. Successively, pairs of AD-Schizophrenia and AD-MS also presented a reasonable number of shared SNPs and genomic loci (Table 1).
| Shared SNPs and Genes Count for 10 pairs of 5 Diseases | ||
|---|---|---|
| Disease Pair | Shared SNP Count | Shared Gene Count |
| AD–PD | 35958 | 1793 |
| AD–T2DM | 2187 | 103 |
| AD–Schizophrenia | 867 | 46 |
| AD–MS | 771 | 62 |
| PD–Schizophrenia | 701 | 24 |
| PD–T2DM | 463 | 21 |
| MS–Schizophrenia | 421 | 28 |
| T2DM–MS | 250 | 17 |
| PD–MS | 246 | 19 |
| T2DM–Schizophrenia | 223 | 18 |
Table 1: List of disease pairs with GWAS associated shared genetic variants and genes count.
The analysis of specific overlapping genome stretches between ‘loci identified for shared GWAS-LD genetic variants’ and ‘loci of already established disease-associated genes in the literature’ revealed that there was a quite significant overlap between GWAS loci and literature based disease-associated gene loci (Supplementary File), which provides suggestive evidence for an association between genetic variants and disease pathology.
Analysis of putative shared pathways was done by mapping genes in genomic hotspots to pathways using MsigDB. Shared pathways-as a functional layer on top of shared genetics - are indicative for putative pathology mechanisms shared between pairs of diseases. The analysis workflow thus identifies disease pairs that do display a high number of shared genomic hotspots, a significant number of putative shared pathways and – as a consequence - may have significantly shared molecular level mechanisms, that-when perturbed - may contribute to disease etiology.
To explore the pathophysiology of putative shared mechanisms in detail, we selected the pair of AD and PD for a mechanistic case study.
Amongst their shared genomic loci, we selected the well-known Tau locus, located on chromosome 17, to explore further detailed molecular mechanisms, as it showed top ranked “functional consequences” scores, based on ENCODE data, the Roadmap Epigenome Consortium data, DNase footprinting analysis, and DNA Methylation data. Apparently, selecting the tau locus seems to add nothing new and novel, as the tau locus is already well known and has been studied in detail. However, this locus has never been studied in a comprehensive way by “embedding” all the affected genes in that locus into a functional context. In our analysis, we expand the mechanistic context associated with the genes in the tau locus, by collecting and assembling all genetic, molecular and statistical evidences from the literature, from patents, from gene expression studies and from knock-out experiments in one comprehensive mechanistic model. In the following, we are presenting this locus in a very novel and unique perspective of stress induced shared pathology of AD and PD.
This genomic hotspot around tau is highlighted in many association studies for multiple statistically significant SNPs (references). The hotspot covers approximately 1 Mb of a chromosomal region characterized by linkage disequilibrium region that contains a large number of genetic variants.
Three genes are prominent in this locus: MAPT (Microtubule- Associated Protein Tau), the CRHR1 receptor-1 (Corticotropin Releasing Hormone Receptor 1) and the CRHR1-IT1 gene (CRHR1 Intronic Transcript 1). These genes are linked to several diseaseassociated genetic markers mapping to both, coding and noncoding regions. Moreover, disease-associated intergenic and intronic SNPs of this locus have several eQTL links with neighboring genes (Supplementary File).
In the course of our investigation of this shared genomic locus, we identified that, other than AD and PD, it also has well-established associations with Stress and Depression phenotypes. We searched for potential genetic, molecular and statistical evidences from the scientific literature and collected additional evidences from patents, gene expression studies and knock-out experiments, that all support the notion of a shared molecular mechanism linking Stress, AD and PD.
To enrich the genetics-driven identification of candidate genes with functional context and to identify potential mechanisms that bear explanatory potential for the presumed shared etiology linked to this particular locus on chromosome 17, we performed a systematic literature analysis using our literature mining environment SCAIView [26]. Contextual information relevant to the previously identified, disease-associated tau locus and being specific for the context of AD and PD was systematically identified and harvested. The extracted information comprises cause-and-effect relationships representing protein-protein interactions, protein inhibitory and activating patterns, protein-complex formation, insights from disease animal model studies, patterns from knockout and gene expression studies, other genetic associations; from gene mapping (fine-mapping) and GWAS meta-analysis studies, and from drug effects; all with high specificity for either AD or PD or both. The vast amount of information extracted was subsequently encoded using the OpenBEL (Open Biological Expression Language) syntax to construct a cause-and-effect computable model [27]. Models were developed separately for human and mouse. The resulting, comprehensive BEL models represent the state of published knowledge in the context of the genes under investigation in the context of AD and PD; the models are then visualized by Cytoscape_v2.8.3 [28] and queried for disease associated molecular mechanisms to unravel mechanistic context that link molecular level perturbation with the disease etiology.
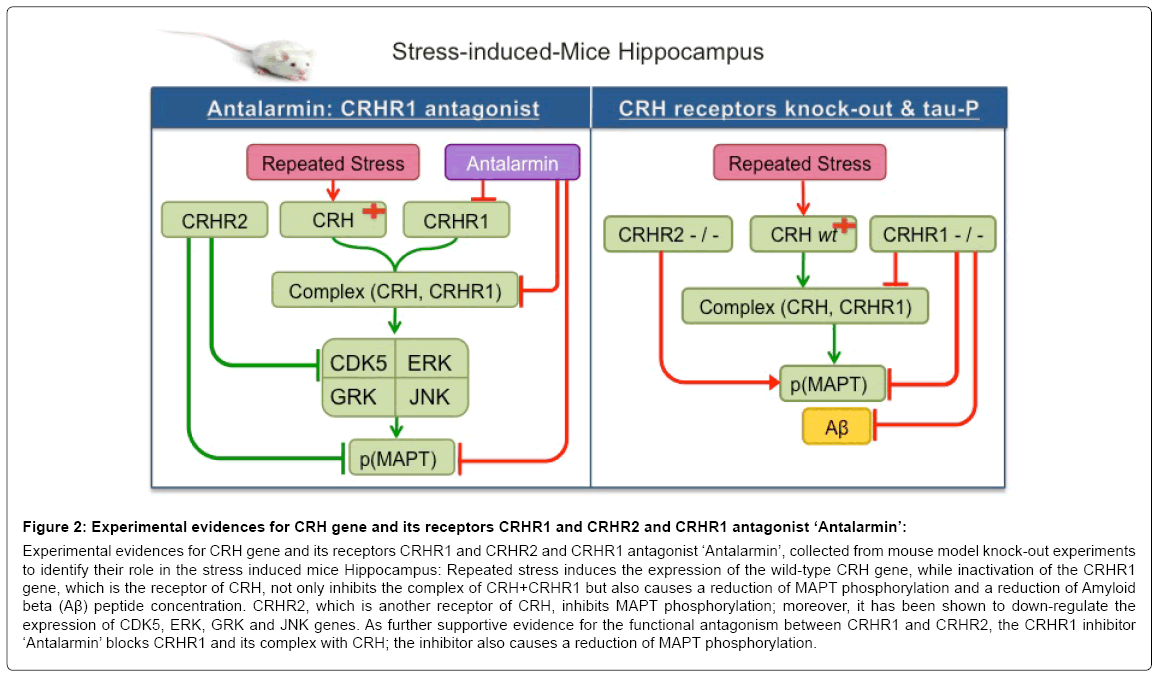
The mouse model reveals that repeated stress induces the expression of the wild-type CRH (Corticotropin Releasing Hormone) gene, while inactivation of the CRHR1 gene, which is the receptor of CRH, not only inhibits the complex of CRH+CRHR1 but also causes a reduction of MAPT phosphorylation and a reduction of Amyloid beta (Aβ) peptide concentration [29]. CRHR2, which is another receptor of CRH, inhibits MAPT phosphorylation; moreover, it has been shown to down-regulate the expression of CDK5, ERK, GRK and JNK genes [30]. As further supportive evidence for the functional antagonism between CRHR1 and CRHR2, the CRHR1 inhibitor ‘Antalarmin’ blocks CRHR1 and its complex with CRH; the inhibitor also causes a reduction of MAPT phosphorylation [30] (Figure 2).
Figure 2: Experimental evidences for CRH gene and its receptors CRHR1 and CRHR2 and CRHR1 antagonist ‘Antalarmin’:
Experimental evidences for CRH gene and its receptors CRHR1 and CRHR2 and CRHR1 antagonist ‘Antalarmin’, collected from mouse model knock-out experiments to identify their role in the stress induced mice Hippocampus: Repeated stress induces the expression of the wild-type CRH gene, while inactivation of the CRHR1 gene, which is the receptor of CRH, not only inhibits the complex of CRH+CRHR1 but also causes a reduction of MAPT phosphorylation and a reduction of Amyloid beta (Aβ) peptide concentration. CRHR2, which is another receptor of CRH, inhibits MAPT phosphorylation; moreover, it has been shown to down-regulate the expression of CDK5, ERK, GRK and JNK genes. As further supportive evidence for the functional antagonism between CRHR1 and CRHR2, the CRHR1 inhibitor ‘Antalarmin’ blocks CRHR1 and its complex with CRH; the inhibitor also causes a reduction of MAPT phosphorylation.
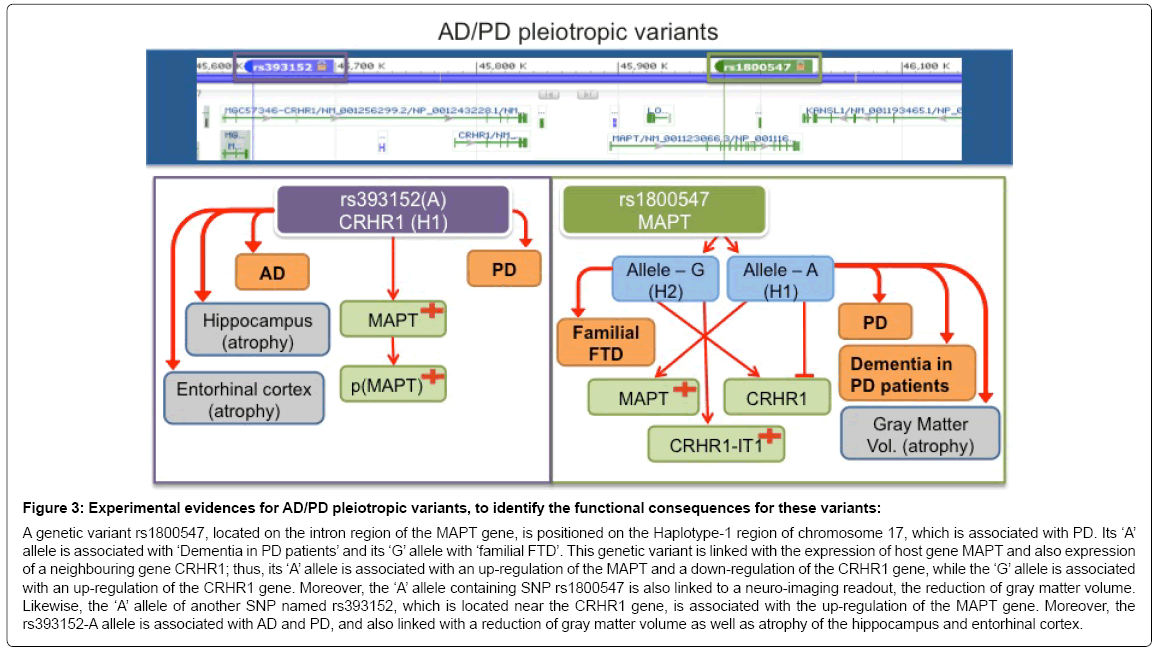
The contextual BEL model specific for human pathophysiology demonstrates that a genetic variant rs1800547, located on the intron region of the MAPT gene, is positioned on the Haplotype-1 region of chromosome 17, which is associated with PD [31,32]. Its ‘A’ allele is associated with ‘Dementia in PD patients’ and its ‘G’ allele with ‘familial FTD’ [33]. This genetic variant is linked with the expression of host gene MAPT and also expression of a neighbouring gene CRHR1; thus, its ‘A’ allele is associated with an up-regulation of MAPT and a concomitant down-regulation of the CRHR1 gene, while the ‘G’ allele is associated with an up-regulation of the CRHR1 gene [33]. In addition, the ‘A’ allele containing SNP rs1800547 is linked to a neuro-imaging readout, the reduction of gray matter volume [33]. Likewise, the ‘A’ allele of another SNP named rs393152, which is located near the CRHR1 gene, is associated with the up-regulation of the MAPT gene [34]. Moreover, the rs393152-A allele has been associated with AD and PD, and seems to be linked to a reduction of gray matter volume as well as atrophy of the hippocampus and entorhinal cortex [34] (Figure 3).
Figure 3: Experimental evidences for AD/PD pleiotropic variants, to identify the functional consequences for these variants:
A genetic variant rs1800547, located on the intron region of the MAPT gene, is positioned on the Haplotype-1 region of chromosome 17, which is associated with PD. Its ‘A’ allele is associated with ‘Dementia in PD patients’ and its ‘G’ allele with ‘familial FTD’. This genetic variant is linked with the expression of host gene MAPT and also expression of a neighbouring gene CRHR1; thus, its ‘A’ allele is associated with an up-regulation of the MAPT and a down-regulation of the CRHR1 gene, while the ‘G’ allele is associated with an up-regulation of the CRHR1 gene. Moreover, the ‘A’ allele containing SNP rs1800547 is also linked to a neuro-imaging readout, the reduction of gray matter volume. Likewise, the ‘A’ allele of another SNP named rs393152, which is located near the CRHR1 gene, is associated with the up-regulation of the MAPT gene. Moreover, the rs393152-A allele is associated with AD and PD, and also linked with a reduction of gray matter volume as well as atrophy of the hippocampus and entorhinal cortex.
BEL models are excellent tools to represent complex physiology; the representation in BEL bears great explanatory potential on how complex physiology works across scales. Our contextual BEL models representing complex physiology of genes in the Tau locus provide a mechanistic explanation, how excessive and repeated stress may modulate pathophysiology. Repeated stress induces the expression of the CRH gene in the hippocampal area [35], while under AD conditions; reduced CRH immune-reactivity is observed [36]. CRH interacts with its receptor, the CRHR1 protein; the CRHR1 gene is highly expressed in hippocampus and the complex between the hormone and its receptor (CRH+CRHR1) can be detected in that brain region [37]. In addition, the CRHR1 protein also interacts with γ-secretase, which is associated with Aβ accumulation, one of the hallmarks of AD pathophysiology [38].
The hormone receptor protein complex (CRH+CRHR1) is further linked to the up-regulation of GSK3β and the phosphorylation of essential elements of the ERK1/2/MAPK pathway [30,39]. Upregulation of GSK3β is associated with MAPT hyper-phosphorylation [30,40]; in addition, phosphorylated MAPT and ERK1/2/MAPK pathway up-regulate Neurofilament phosphorylation, which has been associated with AD [30,39]. The complex physiology is even increased through the interaction of the ‘CRH+CRHR1’ protein complex with the BDNF protein; this interaction has already been associated with AD pathology [41]. The complex also enhances neuronal activity by interacting with adenylate cyclase, cAMP, act(PAK), Ca2 signaling pathways [41]. The resulting enhanced neuronal activity has been shown to further accumulate interstitial fluid amyloid beta (ISF Aβ), while this accumulation of ISF Aβ is also linked with up-regulation of CRH gene expression [42], effectively establishing a feedback loop that can enhance negative dysregulation events. MAPT hyper-phosphorylation also increases its dissociation from microtubules, a process that has been linked to lewy-bodies and Parkinsonism, in the PD context [43].
Finally, the CRHR1 antagonist ‘Antalarmin’, which is used in response of chronic stress, has been shown to reduce Aβ accumulation in brain [44] (Figure 4), adding further meaningful, supportive evidence in context.
Figure 4: Stress induced comorbidity association of HYPERLINK “http://topics.sciencedirect.com/topics/page/Alzheimer’s_disease” AD and PD by genetic variants of Tau locus genes:
Additionally, exhaustive analysis of relevant patent literature revealed that an antagonist against the CRH receptor is effective as a prophylactic or therapeutic agent for diseases, like, anxiety, depression, AD and PD [45]. Patents also describe multiple lines of evidence that suggest the significant role of CRHR1 on neuropsychiatric disorders, and MAPT gene as a well-studied candidate gene for neuropsychiatric disorders [46]. Moreover, it is also patented that although the two receptors CRHR1 and CRHR2 share 70% sequence identity, they differ substantially in ligand binding affinity, and the CRH gene itself has a much higher affinity for CRHR1 rather than CRHR2 [47]. Another patent describes that the accumulation of hyperphosphorylated tau protein in the central nervous system, may be reduced through the administration of CRHR1 selective antagonists and/or CRHR2 selective agonists. Patent [EP2522351 A1] indeed describes methods for the prevention of the onset of Alzheimer’s disease by the administration of CRHR1 selective antagonists [48] (Supplementary File).
Discussion
In the work presented here, we established an integrative approach that starts with a data-driven approach, identifies signals in GWAS data, and gains explanatory potential and allows for new insights into putative complex mechanisms through knowledge-driven context enrichment. Our approach goes way beyond classical “pathway enrichment” approaches, as it takes multimodal information into account and integrates heterogeneous information and knowledge in biologically meaningful, computable graph models. Data, a priori knowledge and inferred insights are combined in a seamless fashion. Meaningful cause-and-effect relationships are established and the signals originally identified are made interpretable in a rational modelling and mining approach.
Our workflow is tailored towards the identification of novel shared mechanisms. It starts with comparative GWAS analysis tailored to identify shared genetic variants; puts the enriched SNPs in contigs (based on linkage disequilibrium); identifies those genes belonging to the “shared” LD loci and establishes compelling evidence for shared molecular mechanisms and biological pathways associated with those genes, for a given pair of disease. The workflow was applied to a comprehensive set of related diseases and allowed us to investigate shared molecular level mechanisms between a pair of diseases, based on both, data driven and knowledge driven strategies.
We would like to emphasize that genomic loci (genomic hotspots) should be considered to investigate the effects of GWAS variants rather than individual genetic variants, particularly to investigate shared pathology. Whereas the biological impact of single SNPs is often hard to predict, the association of several SNPs in a disease-associated LD block provides evidence for a much stronger association that may affect an entire locus with several genes. As a consequence, a set of SNPs in a genomic hotspot may contribute to dysregulation events involving several genes.
Modelling the functional context of these genes in computable cause-and-effect models can be very helpful to identify possible molecular level perturbation mechanisms that contribute to disease pathology. As such, computable mechanistic models are essential to integrate diverse types of data as well as relationships between the nodes; they can help to discover unknown links to illustrate the possible mechanism of dysregulation.
At this point we would like to stress that we are not talking about pathways when we talk about mechanisms. Although pathways are abstractions of biological functional context that is shared by many cell types and often conserved across species boundaries, the pathway concept as it was established over the last 30 years is not taking into account genetic variation information and is not well-suited to take into account the specifics of cell-cell-interactions. “Chains of causation” as we find them in the BEL model graphs may as well exist in pathways, but in pathways they are confined to one type (one “mode” or “level”) of information. Integrative models based on causal relationships, however, span over multiple levels and scales and establish links e.g. from SNPs to imaging features in one single, computable graph model. We would encourage the community to clearly distinguish between pathways (representing canonical information) and mechanisms (representing causes and effects associated with a disease context). Mechanistic modeling allows us to be highly specific with respect to the available knowledge in a given context, without restricting us to make use of canonical knowledge if we wish to include that type of common information.
The mechanistic hypothesis generated from our ‘tau locus BEL model’ establishes a rational, how stress could cause deficits in memory [49-54]. We may actually have established a functional context that puts a “sensor” for environmental and life style into a pathophysiology mechanism that could play a significant role in the etiology of Alzheimer’s disease. Our model provides also mechanistic clue, how hippocampal atrophy may be linked to the pathophysiology of stress [49,51-54] The stress-related HPA axis activation (linked to the CRH-CRHR1 complex) may thus represent a pathophysiological initiation of memory loss [52]. Likewise, it is reported that the decline in CRH Immuno-Reactivity (CRH-IR) in AD is due to the reciprocal accumulation of CRH receptors in affected cortical areas [37]. The alteration in pre- and postsynaptic indicators for CRH is significantly correlated with decline in ChAT (choline acetyltransferase) activity [37].
The H1 haplotype of MAPT extends towards the 5’ region and includes the contiguous gene CRHR1. Linkage Disequilibrium (LD) of this region is substantially associated with PD patients [55]. Strikingly, the oldest and most extensively case-control studies for PD demonstrated the greatest evidence for MAPT and H1 haplotype association. By genotyping H1 haplotype SNPs within the CRHR1- MAPT interval, we can hypothesize that the CRHR1 gene may be responsible for at least part of the disease association of this locus due to the genetic variability and could become a good biomarker candidate, since it is significantly involved in both, immune and nervous systems physiology [55]. Missense and splicing genetic variants in MAPT were first uncovered in ‘frontotemporal dementia with parkinsonism’ associated with chromosome 17 (FTDP-17) [31].
Thus, forgoing studies have already specified associative links between stress, CRH-CRHR1, and tau pathology mediated by CRHCRHR1 dependent activation of tau kinases induced by stress [30,41,56]. On the other side, the H1 haplotype is associated with the accumulation of hyperphosphorylated Tau in neuronal cell bodies, which has always been associated with neurodegenerative diseases [57,58].
Though AD and PD are likely to have different mechanisms underlying their etiology and may affect different brain regions, and display different clinical features, still they have a significant overlap in the progression of neurodegenerative processes. A recent study has been investigating AD and PD GWAS SNPs to identify ADPD pleiotropic genetic variants/loci. They found, that the ‘A’ allele of rs393152, within the CRHR1 and CRHR1-IT1 region (MAPT locus) on chromosome 17, with a MAF (minor allele frequency) value of 23.1%, significantly increased AD and PD risk, additionally, that allele is linked to the up-regulation of MAPT expression [33]. With APOE-stratified GWAS, another study revealed that genetic variants in the chromosome 17q21.31 region are associated with AD [59].
Besides all the genetic evidences that support our mechanistic model, there is also evidence from pharmacology that adds to the plausibility of the pathophysiological context we have established. Rissman et al. described that the selective CRHR1 blocker “antalarmin” blocks stress-induced escalations in tau phosphorylation. This points at a direct function for CRF-dependent signaling in the stress response [30]. Fully in line with this observation is the finding, that CRHR1 antagonist antalarmin is able to suppress amyloid beta accumulation associated with AD pathology, in mice [60].
It is also notable that CRHR1 has a vital role in inflammation [61,62] and the CRHR1 antagonists that are used to treat depression [63,64], also control peripheral inflammation [65-67]. Similarly, those antidepressants, which are known to modulate inflammatory responses, also confer protection against cytokine-induced depressivelike behavioral and biological modifications [68-72].
In a clinical study with MCI (Mild Cognitive Impairment), AD and control groups, Arsenault-Lapierre et al. [73] couldn’t find group differences in cortisol levels. This contradicts several previous studies that found different cortisol levels between normal and AD [74-78] and between normal and MCI groups [78,79]. Contrarily, it supports the literature that found no correlation between cortisol level and perceived stress in different populations [80-82]. Whereas, in this study more MCIs were diabetic, and diabetic patients have been found to secrete higher cortisol levels [83-85], likewise more AD patients were on sedatives or antidepressants. These medications may affect levels of cortisol and perceived stress measured in the patients [73]. However, these findings support our hypotheses that with the mechanism introduced here and the link between HPA axis, genetics and major determinants of AD and PD pathophysiology, we see a source for the highly stochastic nature of sporadic NDDs.
Repetitively, in a recent publication, Park HJ stated that stress response meditated by CRH-CRHR1 mechanism could also contribute to AD pathogenesis [38]. But he also described that under some circumstances, CRHR1 antagonism does not achieve required results against acute stress-induced Aβ production, rather he suggested that either direct targeting of CRH or G protein-biased CRHR1 agonist that could suppress β-arrestin recruitment to CRHR1 might be required to effectively target associated pathway for therapeutic benefit in AD [38].
Conclusion
Our work has established an integrative approach that gains explanatory potential and allows for new insights into putative complex mechanisms through both, data-driven and knowledge-driven context enrichment. Our workflow is tailored towards the identification of novel shared mechanisms and established compelling evidence for a new, shared molecular pathophysiology mechanism associated with AD and PD.
Starting with signal detection in a GWAS meta-analysis, our approach integrates heterogeneous information and knowledge in biologically meaningful and computable graph models. The computable mechanistic model we generated integrates diverse types of data as well as relationships between the nodes, resulting in an efficient ‘functional context enrichment’. We demonstrate that this functional enrichment of genetic variants can lead to the identification of new candidate mechanisms explaining a putative shared aetiology of a given pair of diseases, in our case Alzheimer´s Disease and Parkinsonism.
Though AD and PD affect different brain regions and display different clinical features, still they have a significant overlap in the progression of neurodegenerative processes and our analysis provides compelling evidence for a shared mechanism linking both clinical syndromes.
We emphasize that genomic loci should be considered to investigate the effects of GWAS variants rather than individual genetic variants, particularly to investigate shared pathology, since the biological impact of individual SNPs is often hard to predict.
Funding
This work was supported by the EU/EFPIA Innovative Medicines Initiative Joint Undertaking under AETIONOMY grant agreement n°115568, resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013) and EFPIA companies’ in kind contribution.
Authors Contributions
EY proposed the idea; MN, EY and MHA designed data analysis process; MN analysed the data, generated the results and wrote the manuscript. MHA read, corrected and improved the manuscript. All authors read and approved the manuscript.
References
- Sivakumaran S, Agakov F, Theodoratou E, Prendergast JG, Zgaga L, et al. (2011) Abundant pleiotropy in human complex diseases and traits. Am J Hum Genet 89: 607-618.
- Rzhetsky A, Wajngurt D, Park N, Zheng T (2007) Probing genetic overlap among complex human phenotypes. Proc Natl Acad Sci U S A 104: 11694-11699.
- Stearns FW (2010) One hundred years of pleiotropy: A retrospective. Genetics 186: 767-773.
- Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW (2013) Pleiotropy in complex traits: Challenges and strategies. Nat Rev Genet 14: 483-495.
- Paaby AB, Rockman MV (2013) The many faces of pleiotropy. Trends Genet 29: 66-73.
- Cotsapas C, Voight BF, Rossin E, Lage K, Neale BM, et al. (2011) Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet 7: e1002254.
- Ellinghaus D, Jostins L, Spain SL, Cortes A, Bethune J, et al. (2016) Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet 48: 510-518.
- Parkes M, Cortes A, van Heel DA, Brown MA (2013) Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet 14: 661-673.
- Cross-Disorder Group of the Psychiatric Genomics Consortium (2013) Identification of risk loci with shared effects on five major psychiatric disorders: A genome-wide analysis. Lancet 381: 1371-1379.
- Fortune MD, Guo H, Burren O, Schofield E, Walker NM, et al. (2015) Statistical co-localization of genetic risk variants for related autoimmune diseases in the context of common controls. Nat Genet 47: 839-846.
- Li L, Ruau DJ, Patel CJ, Weber SC, Chen R, et al. (2014) Disease risk factors identified through shared genetic architecture and electronic medical records. Sci Transl Med 6: 234ra57.
- Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, et al. (2013) Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol 31: 1102-1110.
- Pickrell JK, Berisa T, Liu JZ, Ségurel L, Tung JY, et al. (2016) Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet 48: 709-717.
- Li P, Nie Y, Yu J (2015) An effective method to identify shared pathways and common factors among neurodegenerative diseases. PLoS ONE 10: e0143045.
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, et al. (2014) The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 42: D1001-1006.
- Willer CJ, Li Y, Abecasis GR (2010) METAL: Fast and efficient meta-analysis of genome-wide association scans. Bioinformatics 26: 2190-2191.
- Bruehl H, Rueger M, Dziobek I, Sweat V, Tirsi A, et al. (2007) Hypothalamic-pituitary-adrenal axis dysregulation and memory impairments in type 2 diabetes. J Clin Endocrinol Metab 92: 2439-2445.
- Ward LD, Kellis M (2012) HaploReg: A resource for exploring chromatin states, conservation and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40: D930-934.
- Yates A, Akanni W, Amode MR, Barrell D, Billis K, et al. (2016) Ensembl 2016. Nucleic Acids Res. 44(Database issue): D710-D716.
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, et al. (2016) The ensembl variant effect predictor. Genome Biol 17: 122.
- Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, et al. (2016) The Ensembl gene annotation system. Database (Oxford) 2016.
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, et al. (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22: 1790-1797.
- Xie D, Boyle AP, Wu L, Zhai J, Kawli T, et al. (2013) Dynamic trans-acting factor co-localization in human cells. Cell 155: 713-724.
- Younesi E, Toldo L, Müller B, Friedrich CM, Novac N, et al. (2012) Mining biomarker information in biomedical literature. BMC Med Inform Decis Mak 12: 148.
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. (2005) Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102: 15545-15550.
- Hofmann-Apitius M, Fluck J, Furlong L, Fornes O, Kolárik C, et al. (2008) Knowledge environments representing molecular entities for the virtual physiological human. Philos Trans A Math Phys Eng Sci 366: 3091-3110.
- Kodamullil AT, Younesi E, Naz M, Bagewadi S, Hofmann-Apitius M (2015) Computable cause-and-effect models of healthy and Alzheimer's disease states and their mechanistic differential analysis. Alzheimers Dement 11: 1329-1339.
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. (2003) Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498-2504.
- Campbell SN, Zhang C, Roe AD, Lee N, Lao KU, et al. (2015) Impact of CRFR1 ablation on amyloid- β production and accumulation in a mouse model of Alzheimer’s disease. J Alzheimers Dis 45: 1175-1184.
- Rissman RA, Lee KF, Vale W, Sawchenko PE (2007) Corticotropin-releasing factor receptors differentially regulate stress-induced tau phosphorylation. J. Neurosci 27: 6552-6562.
- Hutton M (2001) Missense and splice site mutations in tau associated with FTDP-17: Multiple pathogenic mechanisms. Neurology 56: S21-25.
- Skipper L, Wilkes K, Toft M, Baker M, Lincoln S, et al. (2004) Linkage disequilibrium and association of MAPT H1 in Parkinson disease. Am J Hum Genet 75: 669-677.
- de Jong S, Chepelev I, Janson E, Strengman E, van den Berg LH, et al. (2012) Common inversion polymorphism at 17q21.31 affects expression of multiple genes in tissue-specific manner. BMC Genomics 13: 458.
- Desikan RS, Schork AJ, Wang Y, Witoelar A, Sharma M, et al. (2015) Genetic overlap between Alzheimer's disease and Parkinson's disease at the MAPT locus. Mol Psychiatry 20: 1588-1595.
- Brunson KL, Avishai-Eliner S, Hatalski CG, Baram TZ (2001) Neurobiology of the stress response early in life: Evolution of a concept and the role of corticotropin releasing hormone. Mol Psychiatry 6: 647-656.
- Behan DP, Heinrichs SC, Troncoso JC, Liu XJ, Kawas CH, et al. (1995) Displacement of corticotropin releasing factor from its binding protein as a possible treatment for Alzheimer's disease. Nature 378: 284-287.
- Behan DP, Heinrichs SC, Troncoso JC, Liu XJ, Kawas CH, et al. (1995) Displacement of corticotropin releasing factor from its binding protein as a possible treatment for Alzheimer's disease. Nature 378: 284-287.
- De Souza EB, Whitehouse PJ, Price DL, Vale WW (1987) Abnormalities in corticotropin-releasing hormone (CRH) in Alzheimer's disease and other human disorders. Ann N Y Acad Sci 512: 237-247.
- Park HJ, Ran Y, Jung JI, Holmes O, Price AR, et al. (2015) The stress response neuropeptide CRF increases amyloid-ß production by regulating γ-secretase activity. EMBO J 34: 1674-1686.
- Refojo D, Echenique C, Müller MB, Reul JM, Deussing JM, et al. (2005) Corticotropin-releasing hormone activates ERK1/2 MAPK in specific brain areas. Proc Natl Acad Sci U S A 102: 6183-6188.
- Gong CX, Iqbal K (2008) Hyperphosphorylation of microtubule-associated protein tau: A promising therapeutic target for Alzheimer disease. Curr Med Chem 15: 2321-2328.
- Le MH, Weissmiller AM, Monte L, Lin PH, Hexom TC, et al. (2016) Functional impact of corticotropin-releasing factor exposure on tau phosphorylation and axon transport. PLoS ONE 11: e0147250.
- Kang JE, Cirrito JR, Dong H, Csernansky JG, Holtzman DM (2007) Acute stress increases interstitial fluid amyloid-beta via corticotropin-releasing factor and neuronal activity. Proc Natl Acad Sci U S A 104: 10673-10678.
- Mietelska-Porowska A, Wasik U, Goras M, Filipek A, Niewiadomska G (2014) Tau protein modifications and interactions: Their role in function and dysfunction. Int J Mol Sci 15: 4671-4713.
- Lane RF, Dacks PA, Shineman DW, Fillit HM (2013) Diverse therapeutic targets and biomarkers for Alzheimer's disease and related dementias: Report on the Alzheimer's Drug Discovery Foundation 2012 International Conference on Alzheimer's Drug Discovery. Alzheimers Res Ther 5: 5.
- Nakazato A, Okubo T, Nozawa D, Yamaguchi M, Tamita T, et al. (2004) Pyrrolopyrimidine and pyrrolopyridine derivatives substituted with cyclic amino group. WO/2004/058767.
- Scherer S, Uddin M (2015) Method of determining disease causality of genome mutations. WO/2015/054777.
- Rissman RA, Lee KF, Vale WW, Sawchenko PE (2012) Methods for treatment and prevention of tauopathies and amyloid beta amyloidosis by modulating CRF receptor signaling. EP2522351 A1.
- Mizoguchi K, Kunishita T, Chui DH, Tabira T (1992) Stress induces neuronal death in the hippocampus of castrated rats. Neurosci Lett 138: 157-160.
- Rachal Pugh C, Fleshner M, Watkins LR, Maier SF, Rudy JW (2001) The immune system and memory consolidation: A role for the cytokine IL-1beta. Neurosci Biobehav Rev 25: 29-41.
- McEwen BS (1999) Stress and hippocampal plasticity. Annu Rev Neurosci 22: 105-122.
- McEwen BS, Sapolsky RM (1995) Stress and cognitive function. Curr Opin Neurobiol 5: 205-216.
- Garcia R (2001) Stress, hippocampal plasticity and spatial learning. Synapse 40: 180-183.
- Joseph R (1999) The neurology of traumatic "dissociative" amnesia: Commentary and literature review. Child Abuse Negl 23: 715-727.
- Webster EL, Torpy DJ, Elenkov IJ, Chrousos GP (1998) Corticotropin-releasing hormone and inflammation. Ann N Y Acad Sci 840: 21-32.
- Rissman RA, Staup MA, Lee AR, Justice NJ, Rice KC, et al. (2012) Corticotropin-releasing factor receptor-dependent effects of repeated stress on tau phosphorylation, solubility and aggregation. Proc Natl Acad Sci U S A 109: 6277-6282.
- Webb A, Miller B, Bonasera S, Boxer A, Karydas A, et al. (2008) Role of the tau gene region chromosome inversion in progressive supranuclear palsy, corticobasal degeneration and related disorders. Arch Neurol 65: 1473-1478.
- Stoothoff WH, Johnson GV (2005) Tau phosphorylation: physiological and pathological consequences. Biochim Biophys Acta 1739: 280-297.
- Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert JC, Chung J, et al. (2016) A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry 21: 108-117.
- Lee KW, Kim JB, Seo JS, Kim TK, Im JY, et al. (2009) Behavioral stress accelerates plaque pathogenesis in the brain of Tg2576 mice via generation of metabolic oxidative stress. J Neurochem 108: 165-175.
- Sternberg EM, Hill JM, Chrousos GP, Kamilaris T, Listwak SJ, et al. (1989) Inflammatory mediator-induced hypothalamic-pituitary-adrenal axis activation is defective in streptococcal cell wall arthritis-susceptible Lewis rats. Proc Natl Acad Sci USA 86: 2374-2378.
- Holsboer F (2003) Corticotropin-releasing hormone modulators and depression. Curr Opin Investig Drugs 4: 46-50.
- Chrousos GP (1995) The hypothalamic-pituitary-adrenal axis and immune-mediated inflammation. N Engl J Med 332: 1351-1362.
- Zobel AW, Nickel T, Künzel HE, Ackl N, Sonntag A, et al. (2000) Effects of the high-affinity corticotropin-releasing hormone receptor 1 antagonist R121919 in major depression: The first 20 patients treated. J Psychiatr Res 34: 171-181.
- Webster EL, Lewis DB, Torpy DJ, Zachman EK, Rice KC, et al. (1996) In vivo and in vitro characterization of antalarmin, a nonpeptide corticotropin-releasing hormone (CRH) receptor antagonist: Suppression of pituitary ACTH release and peripheral inflammation. Endocrinology 137: 5450-5747.
- Webster EL, Barrientos RM, Contoreggi C, Isaac MG, Ligier S, et al. (2002) Corticotropin releasing hormone (CRH) antagonist attenuates adjuvant induced arthritis: Role of CRH in peripheral inflammation. J Rheumatol 29: 1252-1261.
- Wlk M, Wang CC, Venihaki M, Liu J, Zhao D, et al. (2002) Corticotropin-releasing hormone antagonists possess anti-inflammatory effects in the mouse ileum. Gastroenterology 123: 505-515.
- Dantzer R, Wollman E, Vitkovic L, Yirmiya R (1999) Cytokines and depression: Fortuitous or causative association? Mol Psychiatry 4: 328-332.
- Licinio J, Wong ML (1999) The role of inflammatory mediators in the biology of major depression: central nervous system cytokines modulate the biological substrate of depressive symptoms, regulate stress-responsive systems and contribute to neurotoxicity and neuroprotection. Mol Psychiatry 4: 317-327.
- Leonard BE (2001) The immune system, depression and the action of antidepressants. Prog Neuropsychopharmacol Biol Psychiatry 25: 767-780.
- Connor TJ, Harkin A, Kelly JP, Leonard BE (2000) Olfactory bulbectomy provokes a suppression of interleukin-1beta and tumour necrosis factor-alpha production in response to an in vivo challenge with lipopolysaccharide: Effect of chronic desipramine treatment. Neuroimmunomodulation 7: 27-35.
- Castanon N, Leonard BE, Neveu PJ, Yirmiya R (2002) Effects of antidepressants on cytokine production and actions. Brain Behav Immun 16: 569-574.
- Arsenault-Lapierre G, Whitehead V, Lupien S, Chertkow H (2012) Effects of anosognosia on perceived stress and cortisol levels in Alzheimer's disease. Int J Alzheimers Dis 2012: 209570.
- Masugi F, Ogihara T, Sakaguchi K, Otsuka A, Tsuchiya Y, et al. (1989) High plasma levels of cortisol in patients with senile dementia of the Alzheimer's type. Methods Find Exp Clin Pharmacol 11: 707-710.
- Davis KL, Davis BM, Greenwald BS, Mohs RC, Mathé AA, et al. (1986) Cortisol and Alzheimer's disease, I: Basal studies. Am J Psychiatry 143: 300-305.
- Maeda K, Tanimoto K, Terada T, Shintani T, Kakigi T (1991) Elevated urinary free cortisol in patients with dementia. Neurobiol Aging 12: 161-163.
- Giubilei F, Patacchioli FR, Antonini G, Sepe Monti M, Tisei P, et al. (2001) Altered circadian cortisol secretion in Alzheimer's disease: Clinical and neuroradiological aspects. J Neurosci Res 66: 262-265.
- Arsenault-Lapierre G, Chertkow H, Lupien S (2010) Seasonal effects on cortisol secretion in normal aging, mild cognitive impairment and Alzheimer's disease. Neurobiol Aging 31: 1051-1054.
- Lind K, Edman A, Nordlund A, Olsson T, Wallin A (2007) Increased saliva cortisol awakening response in patients with mild cognitive impairment. Dement Geriatr Cogn Disord 24: 389-395.
- van Eck M, Berkhof H, Nicolson N, Sulon J (1996) The effects of perceived stress, traits, mood states and stressful daily events on salivary cortisol. Psychosom Med 58: 447-458.
- Murphy L, Denis R, Ward CP, Tartar JL (2010) Academic stresses differentially influences perceived stress, salivary cortisol and immunoglobulin-A in undergraduate students. Stress 13: 365-370.
- Garner B, Phassouliotis C, Phillips LJ, Markulev C, Butselaar F, et al. (2011) Cortisol and dehydroepiandrosterone-sulphate levels correlate with symptom severity in first-episode psychosis. J Psychiatr Res 45: 249-255.
- Lee ZS, Chan JC, Yeung VT, Chow CC, Lau MS, et al. (1999) Plasma insulin, growth hormone, cortisol and central obesity among young Chinese type 2 diabetic patients. Diabetes Care 22: 1450-1457.
- Chiodini I, Di Lembo S, Morelli V, Epaminonda P, Coletti F, et al. (2006) Hypothalamic-pituitary-adrenal activity in type 2 diabetes mellitus: Role of autonomic imbalance. Metabolism 55: 1135-1140.
Citation: Naz M, Younesi E, Hofmann-Apitius M (2017) Systematic Analysis of GWAS Data Reveals Genomic Hotspots for Shared Mechanisms between Neurodegenerative Diseases. J Alzheimers Dis Parkinsonism 7: 368. DOI: 10.4172/2161-0460.1000368
Copyright: © 2017 Naz M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 5700
- [From(publication date): 0-2017 - Apr 02, 2025]
- Breakdown by view type
- HTML page views: 4872
- PDF downloads: 828