Make the best use of Scientific Research and information from our 700+ peer reviewed, Open Access Journals that operates with the help of 50,000+ Editorial Board Members and esteemed reviewers and 1000+ Scientific associations in Medical, Clinical, Pharmaceutical, Engineering, Technology and Management Fields.
Meet Inspiring Speakers and Experts at our 3000+ Global Conferenceseries Events with over 600+ Conferences, 1200+ Symposiums and 1200+ Workshops on Medical, Pharma, Engineering, Science, Technology and Business
Editorial Open Access
System Approach to Analyze High-Throughput Data
| Najafi M1*, Golpoor P1 and Soltanmohammadi E2 | |
| 1Biochemistry Department, Iran University of Medical Sciences, Iran | |
| 2School of Medicine-International Branch, Iran University of Medical Sciences, Iran | |
| Corresponding Author : | Mohammad Najafi Biochemistry Department Iran University of Medical Sciences, Iran Tel: 86703107 E-mail: nbsmmsbn@iums.ac.ir |
| Received: November 16, 2015; Accepted: November 17, 2015; Published: November 24, 2015 | |
| Citation: Najafi M, Golpoor P, Soltanmohammadi E (2015) System Approach to Analyze High-Throughput Data. Biochem Physiol 4:e143. doi:10.4172/2168-9652.1000e143 | |
| Copyright: © 2015 Najafi M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Biochemistry & Physiology: Open Access
| Systems biology is widely applied in translational biomedicine in order to understand and analyze the gene behavior in cellular models over the known biologic pathways. The field is mainly devoted to coordinate the simulation and biological efforts on challenging with bioinformatics and other related areas (OMICS). The main goals of systems biology lie on the clustering of gene profiles, targeting of ligands, integrating and developing of biologic networks by analyzing high-throughput data. Furthermore, it studies the global behavior of components rather than their individual characteristics [1]. |
| Annotation |
| The experimental results obtained in each investigation could be enriched with other data. There are many databases that prepare these data on the basis of genome, transcription and proteome studies. The data can also be obtained from the bioinformatics databases that prepare the molecular and cellular characteristics of network components. Some of databases most used to annotate in systems biology are reported in Table 1. BioMart (biomart.org) and 12D (ophid.utoronto.ca) are other assembled databases that might be used in the enrichment of results. The efforts on the enrichment quality are progressively managed by plugins on networks. |
| Clustering |
| Several clustering models are presented and applied to construct the biologic networks. A network refers to a collection of nodes contacted to edges. Basically, the models are able to build the network blocks, known as hubs. Each hub presents the local contacts of several nodes on the network. Although the hubs obtain the smallest operation units and may indicate the network functional or structural parts, other inter-hub coordinators are the most important linkers to build the network global motifs and modules. The data networking may manually be constructed after annotating the data. However, network analysis depends on the data integration, topology properties and organization characteristics [2]. |
| Method |
| Topological characteristics including network centrality and connectivity are resulted on the modeling techniques. Commonly, two computational methods, known as top-down and bottom-down models, are most used in systems biology. The large-scale data obtained from high-throughput techniques including the protein and gene expression results are generally clustered with the top-down model. On the other hand, the bottom-down model is applied to relatively small data. It is obvious that the use of clustering approaches such as K-means and MCL depends on the primary data and the system understanding of study. |
| Enrichment |
| The networks can be improved by apps in the viewer tools (Cytoscape and String). The network components including nodes and edges may be enriched manually by bioinformatics databases (such as BioMart) or automatically by the compatible plugins. The network processing by annotating data can clear the understanding of disease mechanism, signaling integration and gene regulatory systems [3]. |
| Biochemical Networks |
| Based on the interest of biochemists and biologists, the systems biology improves understanding of cellular events by the system-wide analyses of proteome, genome and transcriptome. This approach opens wide areas to study the molecular interaction, gene regulatory, cell signaling, disease mechanism and response networks. Figure 1 show a protein-protein interaction (PPI) network obtained from gene coexpression data (GEO; GDS3973, DU145 cell line). |
| Perspectives |
| Although systems biology is improving our understanding of disease mechanisms, treatment responses and signaling integrations but its application depends on the advances of tools and bioinformatics. The main challenges can be considered on the scoring approaches of OMICS data, most described as interactome, and computational tools to annotate the network components. Furthermore, the improvement of clustering methods can effectively improve the global network construction [5]. |
References
|
Tables and Figures at a glance
| Table 1 |
Figures at a glance
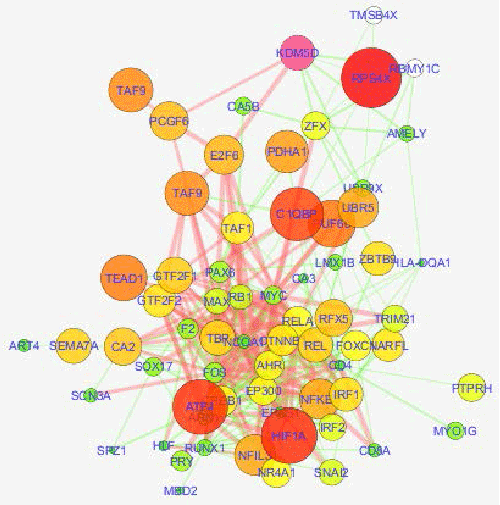 |
| Figure 1 |
Post your comment
Relevant Topics
- Analytical Biochemistry
- Applied Biochemistry
- Carbohydrate Biochemistry
- Cellular Biochemistry
- Clinical_Biochemistry
- Comparative Biochemistry
- Environmental Biochemistry
- Forensic Biochemistry
- Lipid Biochemistry
- Medical_Biochemistry
- Metabolomics
- Nutritional Biochemistry
- Pesticide Biochemistry
- Process Biochemistry
- Protein_Biochemistry
- Single-Cell Biochemistry
- Soil_Biochemistry
Recommended Journals
- Biosensor Journals
- Cellular Biology Journal
- Journal of Biochemistry and Microbial Toxicology
- Journal of Biochemistry and Cell Biology
- Journal of Biological and Medical Sciences
- Journal of Cell Biology & Immunology
- Journal of Cellular and Molecular Pharmacology
- Journal of Chemical Biology & Therapeutics
- Journal of Phytochemicistry And Biochemistry
Article Tools
Article Usage
- Total views: 10009
- [From(publication date):
December-2015 - Jul 18, 2025] - Breakdown by view type
- HTML page views : 9103
- PDF downloads : 906
