Synthesis of a Yeast-Derived Collagen
Received: 09-Aug-2022 / Manuscript No. JBTBM-22-71490 / Editor assigned: 11-Aug-2022 / PreQC No. JBTBM-22-71490(PQ) / Reviewed: 25-Aug-2022 / QC No. JBTBM-22-71490 / Revised: 29-Aug-2022 / Manuscript No. JBTBM-22-71490(R) / Published Date: 05-Sep-2022 DOI: 10.4172/2155-952X.1000290
Abstract
Collagen is a triple-helical mammalian protein, prolific due to its high tensile strength. Collagen’s properties give it applications spanning cosmetics to biomaterials for regenerative medicine. Most collagen is harvested from bovine sources, but this derivation process is unethical and poses health concerns in arsenic poisoning and zoonotic diseases. We developed a new process of generating functional collagen-like protein. Scl, streptococcal collagen-like protein, is a bacterial protein that emulates mammalian collagen properties. The distinction between bacterial and mammalian collagen is the hydroxyproline post-translational mechanisms. Mammalian collagen requires this complex process for high tensile strength, whereas bacterial collagen does not. Thus, Scl2 possesses superiority in mass production while maintaining the qualities of a good biomaterial. Our results include DNA and protein gel verification of our Gibson assembly and lithium acetate transformation into yeast. We also present the results of extracting animal collagen to work on our proposed implementation of collagen hydrogels
Keywords
Biopolymer; Collagen; Scl2
Introduction
From cosmetics to supplements to prosthetics, collagen has countless applications and is prevalent in various products. As the most prevalent protein in the human body, it is in bones, connective tissue, ligaments, and the skin. It is a hard, fibrous, and resistant protein thanks to its triple-helical structure [1]. These properties make it a great biomaterial, applicable as skin meshes, bone grafts, and beauty appliances. It has also seen exceptional results in the meat, food, and beverage industries [2]. In the human body, collagen protein is the building block for wound healing. On damaged connective tissues, collagen folds into a natural scaffold-like shape that is completely nontoxic and biodegradable. It also attracts fibroblasts, which assist in creating scars to cover the wounds [3]. Due to its ability to create new connective tissues to cover wounded skin, scientists have taken steps to create surgical biomaterials.
Current methods of collagen extraction are problematic from an ethical perspective as they involve harvesting calf tendons and hides for collagen samples. Alternative production methods are shown to have dramatically lower yields or are much less efficient therefore, more expensive. While the highest yields of collagen may come from animals, the derivation process is unethical and immoral; said animals (mostly cows) are treated inhumanely and raised only to be slaughtered [4]. Additionally, the byproducts
Bovine collagen harvesting poses significant health concerns to humans. The Food and Drug Administration has found that-due to contaminants-several collagen materials have been the cause of serious problems, such as arsenic poisoning and mad cow disease [5]. These contaminations often occur in slaughterhouses: breeding grounds for zoonotic diseases. There have also been numerous discussions regarding livestock’s effect on the environment. The amount of space, the amount of food and energy, and the resources necessary to grow livestock are taxing on the environment and terribly inefficient. Producing in a lab setting is cheaper and more compact. To bypass the ethical, medical, and financial concerns, we propose expressing collagen through yeast.
We needed to find a way to produce collagen efficiently and ethically, which would be through recombinant expression in E.coli or yeast. There also needed to be a balance on how affordably the collagen could be extracted, so after reading through existing literature, we decided that yeast would be the perfect balance between monetary accessibility and efficiency. To find alternative methods of yielding larger quantities of collagen, we considered multiple bacterial collagen-like proteins as well, such as Bcl (Bacillus colla-gen-like), Pcl (Pneumococcal collagenlike), Buck (Burkholderia collagen-like), and Scl (Streptococcal collagenlike) proteins [6]. Of these, we chose to work with Scl, as it is the most well-known and well-documented collagen-like protein (though still relatively new, allowing for creativity and novelty on our part). Our extraction method uses a Scl2 expression gene and expresses it via S. cerevisiae, a type of yeast. Scl2 has demonstrated qualities similar to collagen with similar triple helical structures - yet with the benefit of not requiring a post-translational modification mechanism (a step necessary for most mammalian collagen). Unlike other approaches to the problem, which also utilize synthetic biology, our yeast-expressed collagen can be used as an engineering template for future research; due to the absence of hydroxyproline and binding sequences, it can be used and tested in many different ways. The novelty of our product lines in its simple versatility: ours is the first to express bacterial collagen in yeast, the first to have gap repair cloning, and since we are not expressing proxyl hydroxylase, we have effectively cut both costs and oxygen.
Numerous biomaterials can be formed using the collagen fiber creation method we propose, from wound dressings, skin grafts, vascular grafts, artificial corneas, and more. The use of collagen in the biomedical industry has endless possibilities, and we hope our protein can act as the base of many more biomaterials.
Methods
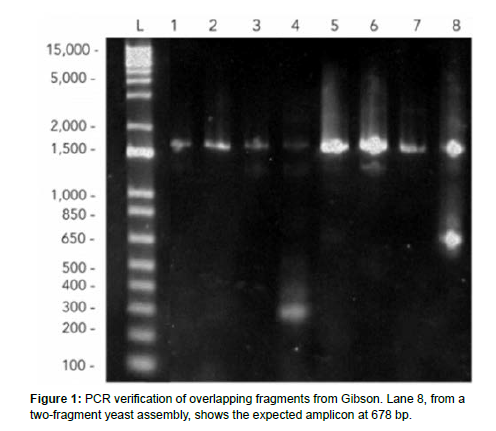
The procedure consists of pelleting yeast cells until suspending them in a LiOAc and DNA mixture in the incubator. Once time has been filled, the yeast can be plated. One yeast assembly was done with two fragments, 3 uL of pRS backbone and 43uL water. This combination ended up being successful, and the yeast plate yielded cells. Next, we grew plates with yeast cultures and used a genomic yeast extraction with ethanol precipitation to break the yeast cell walls to do gel electrophoresis. Then, we picked colonies from the yeast plates to run PCR on overhangs to verify if the transformation worked. Next, we extracted yeast cells from the plates that grew for two days following the transformation to do a yeast miniprep and gel electrophoresis. At first, no bands were visible when run in a gel, which upon reflection may be due to an intact cell wall and failure of the genomic extraction rather than the assembly itself, as we used a different protocol for our last trial that showed successful bands. Later, in the gel electrophoresis, the overhangs were successfully amplified; the correct length on the gel, and the transformed bacterial and yeast colonies with the assembled plasmid grew successfully on both Amp+ and Ura- plates, so the assembly was successful. An amplicon around 678 bp is shown, so we proceeded in our attempts to verify the true expression of the scl2 collagen protein through an SDS-PAGE.
The verification that the DNA had been transformed into the chassis after they grew on selective media was done in several sequential steps. First, for Gibson assembly, the plasmid was extracted via miniprep and run in a gel alongside a linearized pRS4116 fragment to test if the plasmid had increased in size. To verify that the construct was correctly inserted, primers were designed to amplify the overlapping regions on both sides of the construct, with PCR products 678 bp and 713 bp in length, respectively. This was run in a gel to test if the overhangs were assembled correctly. Finally, to verify the expression of the actual protein, several SDS-PAGE trials were run to varying degrees of success, obtaining proper cell wall digestion with Zymolyase to isolate protein lysates and correct staining of the gel with Coomassie.
Results and Discussion
Overview of the Study: The process of synthesizing Streptococcal collagen-like protein, Scl2, was carried out using the complete cds sequence of 1335 bp encoding Scl2 bacterial collagen. Scl2 is encoded in multiple necessary parts; the globular V domain, triple-helical CL domain, and the tail. This insert gene was further modified by adding a 5’UTR upstream of the codon to ensure the triple helical structure of Scl2 is produced. Even further consideration was taken into the promoter and terminator design for the plasmid construct, as specificity is necessary for expressing the bacterial protein in eukaryotic organisms. Plasmid constructs with an optimized terminator reach up to 3.5x higher expression, as found by Curran et al. [7]. The designs were the PRM9 terminator [8], functional naturally in yeast, and the TEF1 pro-moter, also localized to S. cerevisiae. TEF1, however, is not a natural promoter, which reduces the risk of off-target binding for the expression of recombinant constructs. Further upstream, there is a 30 bp space before an FEC activator for the TEF1 promoter. The insert DNA contains the necessary promoter and terminator, 5’UTR, Globular V domain, and Collagen-like Scl2 domain needed to express Scl2.
pRS418 was the backbone utilized in transformation. Before the transformation, the vector was shortened via select PCR amplification using primers. The LacZ gene and f1 ORI were excluded from the replicated region of the backbone. Shortened plasmids have higher efficiency in protein expression and transformation success.
Due to the repeating nature of GLY-XAA-YAA polypeptide sequences in the Scl2 triple-helical CL domain, repeating 80 times, DNA synthesis of the entire Scl2 sequence is challenging and prone to error. Therefore, the inserted gene was synthesized in three different fragments. Fragment 1 contained a block, cpTEF promoter, and 5’UTR. Fragment 2 contained Scl2 encoding DNA. Finally, fragment 3 finished with the PRM9 terminator.
For the process of transforming Scl2 DNA into a highly efficient industrial host, Saccha-romyces cerevisiae was utilized based on its production rates. Specifically, the W303 strain of yeast. The W303 strain used in experimentation contains a URA3 mutation for the purpose of using uracil in yeast plates as a control method for successful transformation. Expressing bacterial collagen in yeast is unprecedented (there has been significant effort in producing human and animal collagen through yeast, but no studies use yeast for bacterial collagen). Still, past studies have shown that the alternative of expressing bacterial collagen in E.coli is problematic, and the yield is often too low.
A yeast assembly, specifically lithium acetate transformation, using the Takara Biosciences procedure was conducted on the fragments of linearized plasmid DNA. Conversely, the linearized components and pRS418 were assembled simultaneously via Gibson assembly. The lithium acetate transformation involved the successful plasmid from Gibson assembly and unlinked linear fragments for gap-repair cloning into S. cerevisiae. As a result, DNA is constructed while preparing for product production and skips the complications of Gibson assembly. There were a total of eight yeast assemblies with LiOAc transformation trials. A standard LiOAc/PEG/H2O protocol was enforced for the first seven trials. Still, the eighth trial of yeast assembly utilized a modified procedure with additional solvents: DMSO and TE buffer. The seventh iteration transformed the successful plasmid from Gibson assembly into yeast and directly transformed linear fragments for gap-repair cloning.
Throughout all of our trials, we recorded the amount of each fragment used in the Gibson Assembly, aiming to use between .2-1.0 pmol as recommended by NEB. Our most suc-cessful trial used the middle range - 0.624 pmol of DNA, with an approximate 2.5:1 ratio of inserts to backbone (Table 1).
| Three-Fragment Construct | Two-Fragment Construct | Three-Fragment Construct | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trial # | 1/3 | 2/3 | 3/3 | pRS4116 | Total | half 2 | pRS4116 | Total | 1_frag | 2_frag | 3_frag | pRS4116 | Total | |
| 1 | 0.266 | 0.362 | 0.218 | 0.090 | 0.935 | 0.350 | 0.256 | 0.250 | 0.856 | 0.198 | 0.219 | 0.231 | 0.140 | 0.788 |
| 2 | 0.266 | 0.265 | 0.093 | 0.624 | 0.296 | 0.340 | 0.231 | 0.175 | 1.042 | |||||
| 3 | 0.266 | 0.265 | 0.093 | 0.624 | 0.166 | 0.370 | 0.343 | 0.135 | 1.014 | |||||
The eighth trial showed the most promising results regarding the number of colonies that grew on selective media. Figure 1 shows that the yeast transformation of the plasmid backbone (as a positive control as it contains a URA3 gene) and Gibson Miniprep grew three colonies, while the negative control had one colony and a two-fragment yeast assembly had no large colonies. The miniprep plasmid from the last successful Gibson showed large colonies.
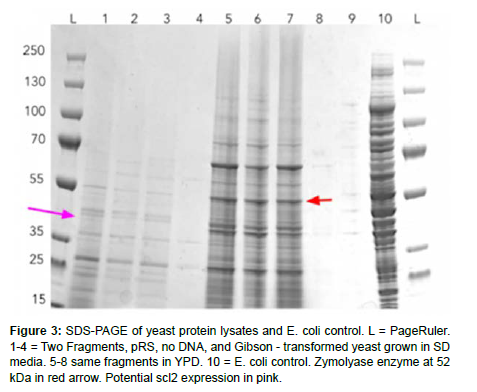
We performed several Gibson assembly trials, lithium acetate transformations, and verification with protein and DNA gels. Although we could not show scl2 expression within our chassis through a clear band on an SDS-PAGE, we could show that our designed plasmid was successfully inserted through colony PCR overhang length verification.
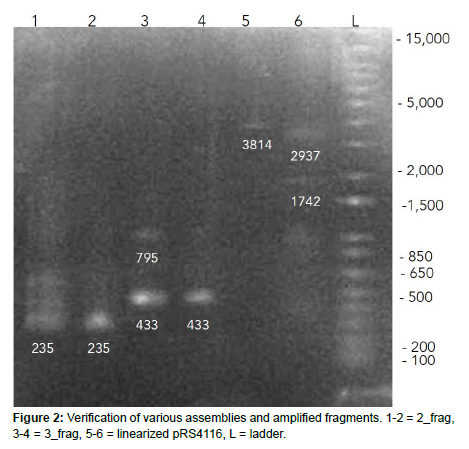
We knew that it would be challenging to express collagen due to the repetitive nature of the collagen-like domain, with 80 Gly-Xaa-Yaa repeats. The countless amount of trials that were required to assemble the plasmid in bacteria and yeast may have been a reflection of this, as working with repetitive sequences is prone to error, especially during PCR which can form hairpin loops and dissociation of the Taq polymerase from the template during amplification, which leads to further “mega primers” that generate unwanted amplicons [9]. As we amplified our gBlocks and genes before Gibson and Yeast assembly, it is possible that the repetitive nature of the scl2 construct prevented precise amplification of our desired sequence. To test this, we ran a gel to verify that our PCR amplicons were present on one of our threefragment designs (Figure 2) The third fragment, which contained the most repetitive sequences, was around our suspected length of 445 bp but included other bands within the lane as expected from mispriming and repetitive sequences.
The final protein gel shows many bands as expected, including the Zymolyase enzyme around 52 kDa, but not the predicted construct around 42 kDa. Nevertheless, we have some promising DNA gel evidence that the construct was successfully inserted into the yeast (Figure 3).
Gel Implementation: The engineering process for implementation was a series of gels testing the effects of different variables and reagents. After eight iterations of testing variables from glycerol to citric acid, we optimized the best gelification formula. Previous iterations had demonstrated instant coagulation when collagen gels were procured in neutral pH rather than acidic pH. Glycerol increased the viscosity of the solution while high collagen concentration predictably bolstered total coagulation but had a noticeable drop-off after 80% concentration.
Our results observed from citric acid and ethanol concentrations had insignificant differences. Still, based on their respective acidic and neutral pH, we speculated that excess of either would result in zero or lots of coagulation. Specifically, too basic or acidic of a pH would instantly dissolve all current coagulation, as observed in the pH trials.
A trend we had noticed was that coagulated collagen would dissipate after a prolonged time. We hypothesized that the disappearance of the coagulation was a result of the supernatant dissolving the bound collagen proteins. Thus, our final iteration added this new consideration on top of the mountains of optimization from previous iterations. We condensed the instantly coagulated collagen and pipetted the supernatant out, replacing it with glutaraldehyde. The result was a small 1cm wide disk of solid collagen gel that acceptably resisted agitation and compression.
The result of our implementation experimentation presents substantial success in the physicality of making a gel. However, it is questionable whether or not the gel we synthesized is indeed made of collagen and whether or not the same process can be recreated with Scl2. Unfortunately, we could not test both questions due to time constraints. But we can confirm that the collagen protein source is undeniably pure, as demonstrated through the SDS pages of the proteins. Thus, our current rationale is that because the collagen source we used was unequivocally collagen protein, the gel is likely to be collagen. This is further supported by our experimentation that showed a proportional relationship between collagen concentration and the amount of coagulation. Furthermore, because Scl2 proteins have the same physical structure, we believe that Scl2 will form a gel with the same gelification procedure.
Thread Implementation: An additional implementation we might explore is that of thread formation. Our primary reference in this avenue is “Collagen Fabrics as Biomaterials,” written in 1994 regarding the derivation of collagen, manufacture of thread, and production of fabrics using unique weaving patterns for the purpose of wound treatment [10].
The procedures of the study, however, must be modified according to the properties and structure of our yeast-expressed collagen and the materials available to us. These modifications might include the following: the development of effective fiber testing procedures; manufacturing methods ensuring the structural stability of the thread; a variation of crosslinking reagents according to final results and overall purpose; altering the wet-spinning apparatus; and varying the collagen source according to respective properties.
The wet-spinning process involves extruding the collagen through a coagulation bath, soaking it in a rinse bath, passing it through an isopropanol bath, drying it under tension in a drying cabinet, and cross-linking it with glutaraldehyde.
Felt Implementation: Another possible implementation of our yeast-expressed collagen is that of felt. For the standard (regular) fabric, its industry-grade manufacture typically involves heat, water, and pressure, with the fibers condensed and pressed together - which permanently interlocks them - creating the matted fabric [11]. Depending on the materials used, the fabric can have different properties, but overall, the most desirable quality of felt is that it involves no knitting or weaving. It serves as one large, flattened piece, which both ensures its structural integrity and greatly minimizes the amount of extra work that goes into production.
Conclusion
The Gibson assembly successfully merged all of our DNA fragments. This is quantifiably visible in the results as the gel electrophoresis of the results of Gibson assembly and PCR indicated the correct bp length around 1500 as well as the expected amplification of extra DNA sequences at 678 bp. Gel electrophoresis of the resultant product of yeast assembly indicated positive results in successful plasmid transformation. Band distribution indicated a significant difference in the control and test groups while the length of the protein expression coincided with previous literature on Scl2 size. The yeastderived collagen has many feasible implementations, which have vast applications. The gel - when further developed - could serve as bioink in the 3D printing of organs, as well as have a wide variety of uses in topical treatments. Collagen thread might be useful in creating sutures, manufacturing woven fabric, or dressing wounds. Felt could also be applied in wound dressing, easier prosthetic integration, and textiles.
Acknowledgements
This work could not have been possible without the support of the Frederick Gardner Cottrell Foundation, which awarded this project the $2500 Impact Grant. Thank you to the San Diego Center for Systems Biology (SDCSB) for granting $2500 to this study. The authors thank Riya I. and Chris J. for their assistance in various tasks such as developing the methods and collecting data. We would also like to thank Dr. Yo Suzuki from the J. Craig Venter Institute for providing materials (including pRS4116 and the W303 yeast strain) and Dr. Peter Zage from the University of California San Diego Moores Cancer Center for his guidance throughout this study.
Competing Interests
The authors declare no competing interests.
References
- Shoulders MD, Raines RT (2009) Collagen Structure and Stability. Annu Rev Biochem 78: 929-958.
- Kolar K (1990) Colorimetric Determination of Hydroxyproline as Measure of Collagen Content in Meat and Meat Products: NMKL Collaborative Study. J Assoc Off Anal Chem 73: 54-57.
- Rangaraj A, Harding KG, Leaper D (2011) Role of collagen in wound management. Wounds UK 7: 54-63.
- Noorzai S, Verbeek CJR, Lay MC, Swan J (2020) Collagen Extraction from Various Waste Bovine Hide Sources. Waste Biomass Valoriza 11: 5687-5698.
- Regenstein JM, Zhou P (2007) Collagen and gelatin from marine by-products. In F. Shahidi (Ed.), Maximising the Value of Marine By-Products. Woodhead Publ pp: 273-303.
- Lukomski S, Bachert BA, Squeglia F, Berisio R (2017) Collagen-like proteins of pathogenic streptococci. Mol Microbiol 103: 919-930.
- Curran KA, Karim AS, Gupta A, Alper HS (2013) Use of expression-enhancing terminators in Saccharomyces cere-visiae to increase mRNA half-life and improve gene expression control for metabolic engineering applications. Metab Eng 19: 88-97.
- Miller C, Schwalb B, Maier K, Schulz D, Dümcke S, et al. (2011) Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol 7: 458.
- Riet J, Ramos LR, Lewis RV, Marins LF (2017) Improving the PCR protocol to amplify a repetitive DNA sequence. Genet Mol Res 16 (3).
- Cavallaro JF, Kemp PD, Kraus KH (1994) Collagen fabrics as biomaterials. Biotechnol Bioeng 43: 781-791.
- Sewport Support Team (2020) What is Felt Fabric: Properties, How its Made and Where. Sewport.
- Atala A, Lanza R, Thomson J, Nerem R (2007) Principles of Regenerative Medicine. Elsevier Science.
- Hayashi T, Nagai Y (1973) Effect of pH on the Stability of Collagen Molecule in Solution. J Biochem 73: 999-1006.
- Ho HO, Lin LH, Sheu MT (1997) Characterization of collagen isolation and application of collagen gel as a drug carrier. J Controlled Release 44: 103-112.
- Hoque A, Mahmood I (2017) Collagen: Changing the Course of the Future. Textile Study Center.
- Lõoke M, Kristjuhan K, Kristjuhan A (2011) Extraction of genomic DNA from yeasts for PCR-based applications. BioTechniques 50: 325-328.
- Osidak EO, Kozhukhov VI, Osidak MS, Domogatskiy SP (2020) Collagen as Bioink for Bioprinting: A Compre-hensive Review. Int J Bioprint 6: 270.
- Peng YY, Howell L, Stoichevska V, Werkmeister JA, Dumsday GJ, et al. (2012) Towards scalable production of a collagen-like protein from Streptococcus pyogenes for biomedical applications. Microb Cell Fact 11: 146.
- Peng YY, Stoichevska V, Madsen S, Howell L, Dumsday GJ, et al. (2014) A simple cost-effective methodology for large-scale purification of recombinant non-animal collagens. Appl Microbiol Bio-technol 98: 1807-1815.
- Ugolini S, Bruschi CV (1996) The red/white colony color assay in the yeast Saccharomyces cerevisiae: epistatic growth advantage of white ade8-18, ade2 cells over red ade2 cells. Curr Genet 30: 485-492.
- Weadock KS, Miller EJ, Bellincampi LD, Zawadsky JP, Dunn MG (1995) Physical crosslinking of collagen fibers: Comparison of ultraviolet irradiation and dehydrothermal treatment. J Biomed Mater Res 29: 1373-1379.
- Xiao T, Staub W, Robles E, Gosse NJ, Cole GJ, et al. (2011) Assembly of Lamina-Specific Neuronal Connections by Slit Bound to Type IV Collagen. Cell 146: 164-176.
- Zhang F, Xie Y, Celik H, Akkus O, Bernacki SH, et al. (2019) Engineering small-caliber vascular grafts from collagen filaments and nanofibers with comparable mechanical properties to native vessels. Biofabrication 11: 035020.
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Citation: Chaturvedi A, Challa A, Zhang N, Thontepu T (2022) Synthesis of a Yeast-Derived Collagen. J Biotechnol Biomater, 12: 289. DOI: 10.4172/2155-952X.1000290
Copyright: © 2022 Chaturvedi A. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 2895
- [From(publication date): 0-2022 - Nov 18, 2025]
- Breakdown by view type
- HTML page views: 2431
- PDF downloads: 464