Research Article Open Access
Structure Analysis of Interacting Domains of RNA Dependent RNA Polymerase (Rdrp) Complex in Nipah Virus
| Md. Jahangir Alam1, Mst. Saleha Sultana1, Jahed Ahmed1*, Auditi Purkaystha1, Abdullah Zubaer2, Mohammad Lashkar2, Kaniz Fatema1 and Md. Rabiul Awal1 | |
| 1Department of Genetic Engineering and Biotechnology, Shahjalal University of Science and Technology, Sylhet-3114, Bangladesh | |
| 2Swapnojaatra Bioresearch Laboratory, Dhaka-1215, Bangladesh | |
| Corresponding Author : | Jahed Ahmed Department of Genetic Engineering and Biotechnology Shahjalal University of Science and Technology Sylhet-3114, Bangladesh Tel: +8801726251159 Fax: +880821-715257 E-mail: jahedahmed@student.sust.edu |
| Received: October 01, 2015; Accepted: October 30, 2015; Published: November 06, 2015 | |
| Citation: Alam MJ, Sultana MS, Ahmed J, Purkaystha A, Zubaer A, et al. (2015) Structure Analysis of Interacting Domains of RNA Dependent RNA Polymerase (Rdrp) Complex in Nipah Virus. Biochem Physiol 4:187. doi:10.4172/2168-9652.1000187 | |
| Copyright: © 2015 Alam MJ, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Biochemistry & Physiology: Open Access
Abstract
Nipah virus is an emerging zoonotic paramyxovirus that is responsible for severe outbreaks in humans and livestocks. This negative stranded RNA virus carries RNA-dependent RNA polymerase (RdRp) complex which is required for viral RNA replication and transcription as a catalytic subunit of viral replicase. We have investigated the 3D structures of four domains from three proteins- Nucleocapsid (N), Phosphoprotein (P) and Polymerase (L) which contribute to form RdRp complex. The presence of intrinsic disorder regions in those proteins under native condition which made the in vitro study of structure difficult. In our study, the 3D homology models of the domain Alpha MoRE and PXD (forming N-P complex), and domain PMD and Domain I (forming P-L complex) were generated and evaluated. Protein-protein docking studies of these four domains was performed which elucidated the structural aspects of RdRp complex and also showed the nature of individual interaction (N-P and P-L). The evidence of the weak binding of Alpha MoRE with PXD than the binding affinity of PMD to Domain I have suggested that, the Alpha MoRE-PXD interaction as a valuable drug target.
| Keywords |
| Nipah Virus; RdRp; PXD; Alpha-MoRE; PMD; Domain-I; Docking |
| Introduction |
| Nipah virus (NiV) is an enveloped negative-stranded RNA virus belonging to the genus Henipa virus of the Paramyxoviridae family. NiV is highly pathogenic and emerging zoonotic virus that is responsible for several outbreaks of severe respiratory illness and viral encephalitis, first recognized in Malaysia on September, 1998 [1,2]. As in all mononegavirales members, the RNA genome of Nipah virus is encapsulated by a viral coded nucleoprotein (N) to form a ribonucleoprotein (RNP) complex. This RNP serves as the template for viral RNA synthesis by the polymerase (L) protein during both transcription and replication [3-7]. The encapsulation provides the way for phosphoprotein to bind with N protein and recruit L protein [8-10]. |
| Cytosolic transcription and replication of the viral genome is mediated by the RNA-dependent RNA polymerase (RdRp) complex, which includes the ribonucleoprotein (RNP) and viral P and L proteins [11-13]. The complex is formed by the domains α-MoRE from N protein, PXD and PMD from P protein [14,15] and Domain I from L protein [16]. The N and P proteins have been shown to interact with each other being able to form Alpha MoRE-PXD (N-P) interaction where the P protein and the L protein form PMD-Domain I (P-L) which latter serve as a way to recruit L onto nucleocapsid template [17]. The two interactions must be formed separately prior to the binding of RdRp complex with RNA template for replication. N-P and P-L is essential for their biological activity in nucleocapsid RNA replication in vitro [18]. |
| Previous studies showed that RNA virus proteins are enriched in intrinsic disordered regions (IDR) due to their high mutation rate [19]. In normal physiological condition IDRs have no define secondary and tertiary structure [20]. The computational and experimental studies proved that MeV proteins involved in replicative complex have large intrinsically disordered regions (IDRs) [21-23]. Using computational approaches, we render the result of disorderness to all proteins in Nipah virus and show the abundance of IDRs within the RdRp complex of the virus. Due to the abundance of disorderness in whole proteins found in RdRp complex, we report the structural prediction of four domains, i. e., Alpha MoRE, PXD, PMD and Domain I which form the complex. In our study, we propose the Alpha MoRE-PXD interaction in RdRp complex as a putative drug target to inhibit the virus. The approach aimed at disrupting this interaction and blocking transcription and replication of virus. |
| Materials and Methods |
| Sequence retrieval and intrinsically disordered location prediction |
| Protein sequences of different domains of RdRp complex were retrieved as FASTA format from NCBI () and ViPR (http://www. viprbrc.org/) for this study. Identification of intrinsically disordered location of these domains was performed by DisEMBL [24], RONN [25] and IUPred [26] prediction program. DisEMBL was run using default settings. The Hot-loop and Coil results were both included in our evaluation. IUPred was run under the long sequence default settings. RONN was used in default setting. Depending on prediction, the results of all amino acids were utilized to a 0-1 scale. Residues with values of 0.5 or greater indicate the possibility to be disordered. According to this analysis values of all residues were further combined and averaged. |
| Secondary structure prediction |
| The secondary structures of four domains were analyzed by using Jpred [27], PORTER [28], and YASPIN [29]. Jpred is followed by combination of three programs (Jnet, Jhmm, Jpssm).All programs were used in default settings providing the result of helix, strand and coil with the confidence value (0-9) where 9 is the best confidence. |
| Homology modelling and evaluation |
| Modeller 9.15 [30] was used for homology modeling of protein three dimensional structures. The multiple alignment of query sequence was provided with known sequences called templates to calculate a 3-D model containing all non-hydrogen bond atoms. In order to confirm suitable templates, PSI-BLAST [31] against all existing molecules in the protein data bank (PDB) was performed. Besides, HHpred [32], ps2v2 [33], phyre2 [34] and CPH models 3.2 server [35] were also used to get suitable template. The ps2v2, phyre2, CPH models 3.2 servers were provided direct model of target domain. The model of Alpha MoRE, PXD, PMD were found from phyre server and the model of Domain I was found from CPH models server which used as template. |
| The stereo-chemical properties of protein models were assessed by two different servers, i.e., RAMPAGE [36] and ERRATv2 [37]. RAMPAGE provides Ramachandran plot to analyse the quality of model where >98% of the favorable region resemble as a good quality model. Good model with high resolution structures generally produce values around or higher than 90% in ERRATv2. |
| Protein-protein docking |
| Molecular docking was performed by two web-based servers Patchdock [38] and pyDock [39] to identify the substrate binding site and affinity of proteins. Patchdock compute the molecular surface of protein and discard the complex with unacceptable penetration of one protein to another protein. Finally it gives complex with ranking according to geometrically shape complementarities scores. pyDock generates the result with best rigid body orientation including electrostatic, desolvation energy and limited vanderwaals contribution. In the analysis, 50 protein-protein complexes were analysed for each interaction from which the best fitting was chosen. The global energy of both two interactions was tested in Firedock [40]. The molecular graphics software PyMOL [41] was used to visualize. The root mean square deviations (RMSD) between the models and the template were calculated by PyMOL. The RMSD value of two interactions found from Patchdock and pyDock were also calculated by the same software. |
| Protein-protein interaction analysis |
| Protein interaction calculator [42] analyses protein-protein interaction by calculating disulphide bonds, hydrophobic interactions, ionic interactions, hydrogen bonds, aromatic-aromatic interactions, aromatic-sulphur interactions and cation-π interactions between proteins in the complex. The Protein Interfaces, Surfaces, and Assemblies (PISA) server [43] was used to analyze the interface in the models of both complexes. |
| Results and Discussion |
| Prediction of intrinsic disorder regions |
| According to structural analysis, intrinsic disorder regions prediction which provides the way to characterize protein with defines secondary and tertiary structure. In the result, ~ 30% amino acid (Table 1) in N protein (532 amino acid residues) has disordered amino acid and most prominently the C-terminal region (after 400 amino acid residues) show greater unstructured amino acid which is the interaction prone area. P protein (Table 1) contains the variable amino acid almost in all over the protein having ~ 50% disorderness. L protein (Table 1) in the same complex show little disorderness. |
| The order structures of L protein have been supposed to bear precise protein scaffold which have enzymatic activities for transcription and replication. The present study has shown that long order structured regions were found in N and P which are essential to form RdRp complex suggesting a strong overall trend of disorder in RdRp complex. As the flexible region has no define secondary and tertiary structure the study has focused on four interacting domain of all three proteins and explored out their ordered structure. |
| Secondary structure analysis and homology modelling |
| As RdRp complex acts as a catalytic subunit of viral replicase, it plays important role in increasing the severity and number of this infectious virus through regulating the replication and transcription of virus itself. So it is essential to predict the tertiary structure of the complex where the secondary structure show the amino acid lies in helix, strand or coil. The result of secondary structure revealed that alpha helix dominating among secondary structure elements followed by alpha helix, extended strand and beta turns for all sequences. In this study all domains show >70% alpha helical structure (Table 2) and lack of beta sheet except PMD domain which have very few amino acid shown beta sheet in prediction. |
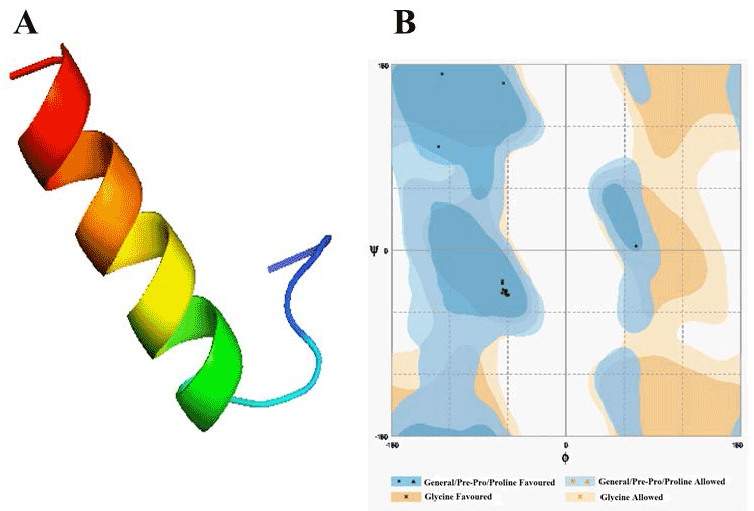
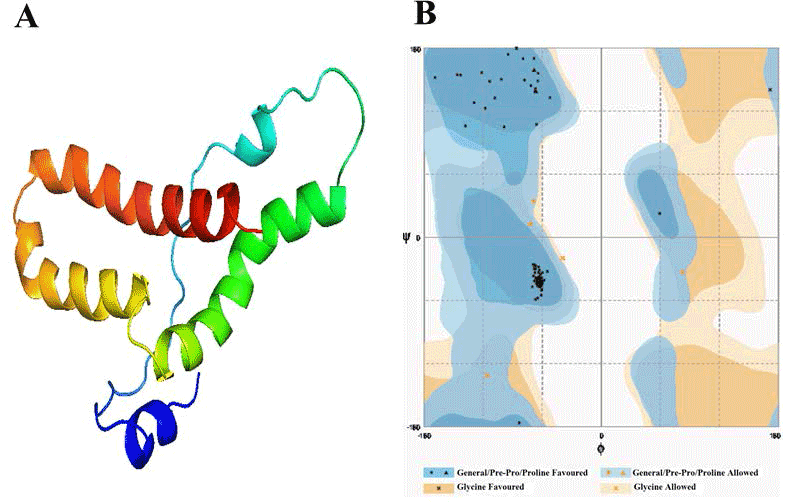
| Template identification server have suggested template for modelling of these domain. Comparative modelling template was unable to give a good model of required domains. Therefore to get a good model, we had used theoretical model as template found from different servers for each domain. In this study, the model of Alpha MoRE, PXD, PMD and the model of domain I were found from phyre server and CPH models server, respectively, were used as template. The result of Alpha MoRE (Figure 1), PXD (Figure 2), PMD (Figure 3) and Domain I (Figure 4) showed 100%, 97.9%, 99.1% and 96.3% respectively in the favorable region in ramachandran plot (Table 3). All these results suggested that all models were reliable and structurally validated. The overall quality factor were investigated by ERRAT (Table 3) which scores about 100%, 97.619%, 98.077% and 92.350% in Alpha MoRE (Figure 1), PXD (Figure 2), PMD (Figure 3) and Domain I (Figure 4), respectively. The predicted structures were confirmed well to the stereochemistry indicating a reasonably good quality model. The quality of a model can also be approximately predicted from the sequence similarity of both structure (model used as template and the final model). The RMSD of four models were 0.703 Å, 0.264 Å, (12.992 & 0.762) Å, 0.722 Å of Alpha MoRE (Figure 1), PXD (Figure 2), PMD (Figure 3), Domain I (Figure 4), respectively, referring the model are similar from the model generated by the server. Therefore, the predicted models were acceptable for further structural analysis. The results of validation suggest the better understanding of conformational figures of all models. These structures will provide a good foundation for structural analysis of total RdRp complex. |
| Protein-protein docking and interaction analysis |
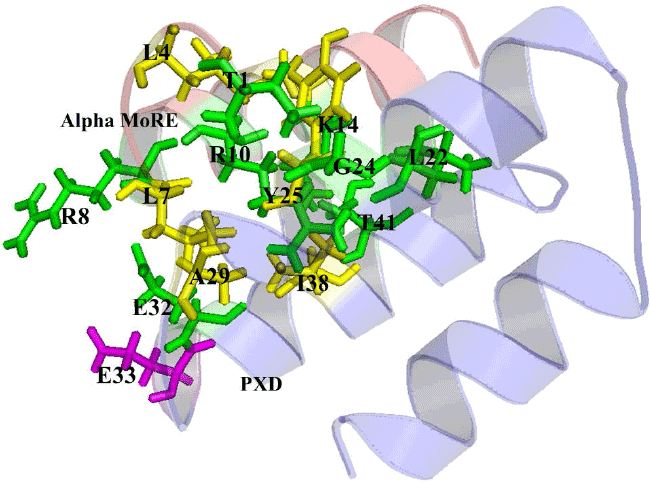
| The analysis of protein interaction with other proteins is the key point to know the intermolecular complexity of any complex formed by two or more proteins. In this study, protein-protein docking was applied to explore the binding mechanism in RdRp complex. The binding analysis provides the overall structural feature of the complex and gives the information about binding affinity of one protein with another. The binding of Alpha MoRE to PXD is shown in Figure 5 which have four hydrophobic interaction with 5Å and four hydrogen bond with 4Å (Table 4). |
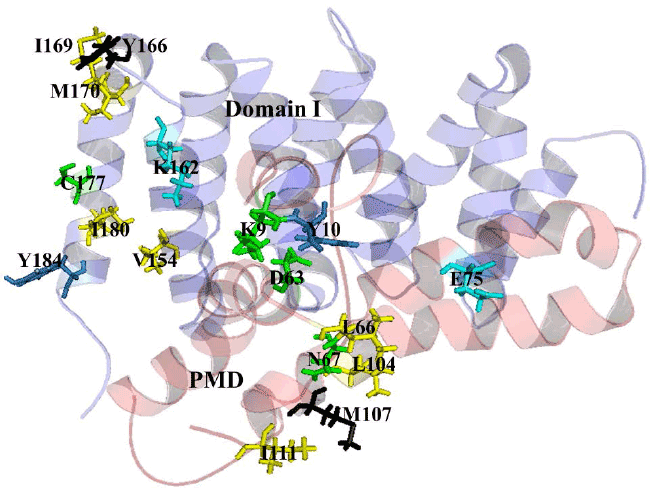
| The pairwise RMSD value calculated by pymol for Alpha MoREPXD interaction is 0.000 in PXD and 0.001 in Alpha MoRE however, the complex from patchdock align with the complex from pyDock referring that the complex has no difference in either server. PMD bind with Domain I (Figure 6) by 10 hydrophobic interactions and 3 hydrogen bonds (Table 5). In the case of PMD-Domain I, the pairwise RMSD value is 0.003 in PMD and 0.000 in Domain I calculated according to the same way that was done Alpha MoRE-PXD interaction. Consequently, both of two interactions have the validity for further analysis. |
| Comparison between two interactions |
| The protein-protein interaction plays an important role in structure based drug target identification. The study opens the way to the structural evaluation of RdRp complex, because the gain of structure arising from two interactions may lead to the formation of the complex. The strongest binding interaction was characterized by protein-protein binding energy which is given in Table 6. The interface areas of 631.4 Å2 and 726.8 Å2 of PXD-Alpha MoRE (Figure 5) and PMD-Domain I (Figure 6) respectively represent that surface buried in PXD by the Alpha MoRE is smaller than the buried surface area of Domain I by PMD domain. |
| It implies that PXD-Alpha MoRE interaction is less strong than PMD-Domain I interaction. This assume due to previous studies which reveal that weak complexes have loosely packed interfaces that are smaller than in tight complexes [44]. The lower buried surface area of the NiV Alpha MoRE-PXD interaction is consistent with the lower affinity of the binding reaction as compared with the PMD-DomainI interaction. Moreover another reason for less tightly bound complex of NiV PXD-Alpha MoRE is the type of interaction and their number. The PXD-Alpha MoRE share 4 hydrophobic interactions where PMDDomain I share 10 hydrophobic interactions suggesting complexity as more hydrophobicity infer stronger binding. Additionally PMD-Domain I have aromatic-aromatic and aromatic-sulpher bond determine the increasing of binding affinity of PMD to Domain I (Table 6). |
| The Alpha MoRE-PXD complex cannot be excessively stable for this transition to occur efficiently at a high rate. By the analysis of the result, it is predicted that the PXD domain of P protein can be used as drug target for protein ligand docking approach. |
| Conclusion |
| We presented the prediction of disorderness in proteins of RdRp complex which have highly disordered region. By this finding we fuel up our work to design the model of interacting domain rather than whole, contribute to form RdRp complex. Tertiary structure of these domains helps us to get the pattern of interaction with one another. It reports that Alpha MoRE-PXD interaction is more flexible than PMD-DomainI interaction. PMD-Domain I move through PXDAlpha MoRE interaction during viral RNA synthesis [14] and from our analysis we have found PXD-Alpha MoRE interaction is less stable than PMD-Domain I interaction considering that the contact between PXD and Alpha MoRE within the RdRp complex has formed/deformed repeatedly to give access the polymerase (L protein) to move through the nucleocapsid template. We concluded our study by providing result that the Alpha MoRE-PXD interaction can be used as a target for antiviral drug design. This study has given an idea to design a new drug molecule against the PXD domain which will bind with more affinity than Alpha MoRE. This inhibition of PXD-Alpha MoRE interaction will stop the formation of RdRp complex hence transcription and replication also be stopped and the virus will be inhibited. |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 |
| Table 4 | Table 5 | Table 6 |
Figures at a glance
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 |
 |
 |
 |
| Figure 4 | Figure 5 | Figure 6 |
Relevant Topics
- Analytical Biochemistry
- Applied Biochemistry
- Carbohydrate Biochemistry
- Cellular Biochemistry
- Clinical_Biochemistry
- Comparative Biochemistry
- Environmental Biochemistry
- Forensic Biochemistry
- Lipid Biochemistry
- Medical_Biochemistry
- Metabolomics
- Nutritional Biochemistry
- Pesticide Biochemistry
- Process Biochemistry
- Protein_Biochemistry
- Single-Cell Biochemistry
- Soil_Biochemistry
Recommended Journals
- Biosensor Journals
- Cellular Biology Journal
- Journal of Biochemistry and Microbial Toxicology
- Journal of Biochemistry and Cell Biology
- Journal of Biological and Medical Sciences
- Journal of Cell Biology & Immunology
- Journal of Cellular and Molecular Pharmacology
- Journal of Chemical Biology & Therapeutics
- Journal of Phytochemicistry And Biochemistry
Article Tools
Article Usage
- Total views: 17820
- [From(publication date):
December-2015 - Nov 24, 2024] - Breakdown by view type
- HTML page views : 13330
- PDF downloads : 4490
