Research Article Open Access
Structural Effects of Leigh Syndrome Mutations on the Function of Human Mitochondrial Complex-I Q module
| Tulika M Jaokar, Ranu Sharma and Suresh CG* | |
| Division of Biochemical Sciences, National Chemical Laboratory, Dr Homi Bhabha Road, Pune-411008, India | |
| *Corresponding Author : | Suresh CG Division of Biochemical Sciences National Chemical Laboratory Dr Homi Bhabha Road, Pune-411008, India Tel: 91-20- 25902236 Fax: 91-20-25902648 E-mail: cg.suresh@ncl.res.in |
| Received January 23, 2013; Accepted March 01, 2013; Published March 04, 2013 | |
| Citation: Jaokar TM, Sharma R, Suresh CG (2013) Structural Effects of Leigh Syndrome Mutations on the Function of Human Mitochondrial Complex-I Q module. Biochem Physiol S2:004. doi:10.4172/2168-9652.S2-004 | |
| Copyright: © 2013 Jaokar TM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Biochemistry & Physiology: Open Access
Abstract
The Q module of the human mitochondrial Complex-I (NADH:ubiquinone oxidoreductase) comprises of four core protein subunits: NDUFS2, NDUFS3, NDUFS7 and NDUFS8. It is an intermediate unit, connecting the dehydrogenase domain (N module) and the membrane arm (P module) of the L shaped mitochondrial Complex-I. Its role is to transfer electrons from N module to the P module. Mutations in the subunits of this module are reported to be associated with Leigh syndrome, a neurological genetic disorder. The Q module of human mitochondrial Complex-I is modelled based on the crystal structure of Thermus thermophilus Complex-I. The structural ramifications of the documented Leigh syndrome mutations were studied in silico, using molecular dynamics simulations. Although the mutations caused minimum perturbation to the overall secondary structure, the root mean square fluctuations of certain segments were substantial, especially those in the loop regions. The mutations affected the hydrogen bonded interactions, solvent accessible area, and the observed radius of gyration to various extents. The cumulative effect of all these changes on the formation of complex assembly is reflected in the observed unfavourable energy variations, instability of subunit association, and a reduced affinity for substrates, such as ubiquinone. Thus, our analysis indicates that Leigh syndrome mutations lead to formation of structurally and functionally defective complex, which in turn results in disease phenotype.
| Keywords |
| N, Q and P module; Homology modelling; Molecular dynamics simulation; Root mean square fluctuation; Radius of gyration; n-decyl-ubiquinone; Binding affinity |
| Abbreviations |
| PDB: Protein databank; NADH: Reduced Nicotinamide Adenine Dinucleotide; FMN: Flavin Mononucleotide; MD: Molecular Dynamics; DBQ: n-decyl-ubiquinone; WT: Wild Type; RMSD: Root Mean Square Deviation; RMSF: Root Mean Square Fluctuation; Rg: Radius of gyration; BN-PAGE: Blue Native Polyacrylamide Gel Electrophoresis |
| Introduction |
| NADH: Ubiquinone Oxidoreductase or Complex-I of the respiratory electron transport chain is the largest multi-subunit enzyme complex, comprising of ~ 14 subunits in prokaryotes and 42 subunits in human mitochondria. Complex-I transfers electrons from NADH to FMN, through a series of eight iron-sulphur centres and finally, to the terminal electron acceptor ubiquinone. It also possesses proton pumping activity. The L-shaped structure of Complex-I is organized into three domains: dehydrogenase domain (N module), hydrogenase domain (Q module), and the membrane arm (P module). The hydrogenase domain or the Q module, also known as ubiquinone reduction module, is made up of four core subunits: NDUFS2, NDUFS3, NDUFS7 and NDUFS8. These subunits encoded by the nuclear genome are a part of the peripheral arm, which connects the dehydrogenase domain to the membrane arm in the L-shaped complex [1], and are involved in electron transfer from N module to membrane arm. The ubiquinone reducing catalytic core is located at the interface of subunits, NDUFS2 and NDUFS7 [2]. Ubiquinone plays a dual role by acting as a substrate, and as a tightly bound co-factor [3]. The Q module harbours three ironsulphur clusters. One of them, cluster N2, acts as an electron donating group. This cluster, located at the end of the redox centres, transfers electrons from the flavin site to quinine [4]. This is also suggested to play a role in coupling the redox reactions that translocates protons across the membrane [5]. Thus, Q module is also the electron donor for the ubiquinone [6,7]. In addition, NDUFS2 harbours binding sites for quinone related Complex-I inhibitors, such as piericidin and rotenone [8]. |
| The core subunits of complex-I show strong conservation of C-terminal sequence across species (Supplementary Figures SF1-SF4). In humans, the mutations in subunits NDUFS2, 3, 7 and 8, are associated with neurological genetic disorders like Leigh syndrome and Leigh-like syndrome [9-16]. Mutations in these subunits are reported to cause functional deficiencies and accumulation of assembly intermediates. Structural and molecular dynamics study of the subunits is expected to provide insights into the assembly of Q module and the role of subunit-mutations in leading to disorders, such as Leigh syndrome. In the present study, the Q module of human mitochondrial Complex-I has been modelled based on the crystal structure of the hydrophilic domain of Thermus thermophilus. Molecular models of individual core subunits were used in molecular dynamics (MD) simulations, and conformational changes on a time scale was monitored to assess the consequences of known mutations on the stability of individual subunits. Molecular modelling and simulation studies of the Q module detected several changes in mutant structures. Functional consequences of the mutations were evaluated by comparing the calculated binding affinities of mutants and wild-type structures for n-decyl-ubiquinone (DBQ), and iron-sulphur clusters. Energy and stability differences between mutants and wild-type subunits were estimated. |
| We have applied in silico modelling combined with molecular dynamics, to elucidate the effects of mutations associated with Leigh or Leigh-like syndrome, and thus, understand their mechanism of disruption of the structure, assembly and function of Complex I Q module. |
| Materials and Methods |
| Molecular modelling of the four protein subunits of Q module |
| The sequences for the four subunits of Q module were taken from the UniProtKB (http://uniprot.org) protein databank (Entry number: O75306, O75489, O75251, O00217). A similarity search carried out using the BLAST server [17], identified in Protein Databank (PDB) (http://www.rcsb.org), the bacterial complex (PDB ID: 2FUG, 3I9V, 3IAM) of the organism Thermus thermophilus, which had 40-50% sequence identity with the corresponding four subunits of human Complex-I Q module [5,18]. The subunits, NDUFS2 and NDUFS7, were modelled using D and F chains of 3I9V (resolution: 3.1 Å, whereas subunits NDUFS3 and NDUFS8, were modelled using E and G chains of 3IAM (3.1 Å). The signal sequences for subunits were predicted using Mitoprot [19], and were excluded from modelling. The molecular models were prepared using the software Modeller9.10 [20]. The in silico mutants were generated using the program FoldX [21]. Energy minimization of both wild type and mutants was by using Gromacs v.4.5 [22], by the steepest descent minimization for 100 ps, with maximum force field cut off being 1 KJ/mol. The molecular models were evaluated using Errat (version 2.0) [23], PDBsum [24], ProSAweb Protein structure analysis [25], and RMSD based on Cα overlap between target and template (Table 1). |
| Molecular dynamics (MD) simulation of subunits |
| The molecular models of wild-type and mutant proteins were used to perform molecular dynamics simulation using Gromacs v.4.5, with the OPLS-AA/L all-atom force field. The subunits were solvated with SPC water model, using the genbox program of Gromacs suite. The cubic boxes chosen default by Gromacs with dimensions: 12.03 nm (NDUFS2), 10.55 nm (NDUFS3), 9.38 nm (NDUFS7) and 7.49 nm (NDUFS8) were used. Sodium and chloride ions were added to each system, depending on requirement for charge neutralization (Supplementary Table 1). Energy minimization was performed by the steepest descent method for 50,000 steps (the minimization tolerance was set to 1000 kJ/mol-nm). Equilibration was carried out in 2 steps, using conditions NVT and NPT, respectively. Long range electrostatics was computed using the Particle Mesh Ewald (PME) method [26], and Lennard-Jones energy cut off was set to 1.0 nm. Bond lengths were constrained with the LINCS algorithm [27]. Simulations of 15 ns duration were performed on the wild type, and mutant structures at constant temperature of 300 K maintained by modified Berendsen thermostat coupling [28], and at a constant pressure of 1 bar by Parrinello-Rahman pressure coupling. The time step employed was 2 fs, and coordinates were saved every 2 ps for analysis of MD trajectory. Analyses were performed with the tools available in the Gromacs utilities. RMSD, RMSF, Radius of gyration (Rg), average number of inter and intra-molecular hydrogen bonds formed, solvent accessible area and secondary structure prediction along the trajectory (DSSP), were estimated. |
| Modelling Q module assembly |
| The subunit assembly of Q module was modelled by comparing with the arrangement of chains D, E, F and G (PDB ID: 2I9V), using the alignment program in PRIME v3.1 (Schrodinger) [29]. The initial model of Q module, along with three iron-sulphur clusters, was first generated. The model was then energy minimized and prepared in the “protein preparation wizard” of Maestro9.3 [30]. The quality of assembly was assessed using Errat (version 2.0), PDBsum, ProSa-overall model quality plot, and RMSD of Cα positions between target and template. |
| Docking of n-decyl-ubiquinone (DBQ) in Q module |
| The molecular model of Q module prepared, as already described, was used to dock n-decyl-ubiquinone (DBQ), using Glide 5.8 (Schrodinger) [31,32]. The model structure was imported and a centroid receptor grid was generated around the residue, Y141, known to be interacting with DBQ [7]. The ligand molecule DBQ (CID 2971) was downloaded from PubChem compounds [33], and prepared in the LigPrep version 2.5 of the Schrodinger suite [34]. This was then docked in the Q module and ten poses were generated. Out of these poses, the most likely one was chosen based on parameters, such as glide gscore, glide emodel score, and essential interactions were confirmed by experimental mutagenesis data [7]. |
| In silico mutation analysis |
| Residue scanning wizard of Bioluminate 1.0 in Schrodinger suite [30], was used to study the structural effects of mutations on the Q module. The difference in the stability of mutant compared to wild-type (WS) protein was estimated for individual mutations. Similarly, the difference in binding affinity of each mutant for the three iron-sulphur clusters, the DBQ, and other interacting subunits in comparison with corresponding wild-type, was also considered. |
| Results |
| Modelled structures of Q module subunits |
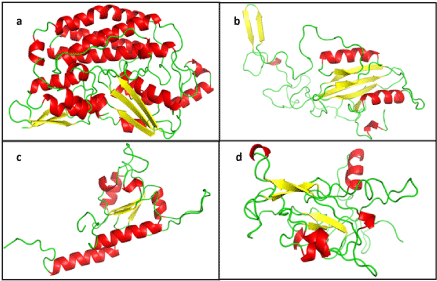
| Ten molecular models were generated for each subunit, using corresponding templates. The signal sequence was not modelled. The N-terminal residues, first 49 of NDUFS2 and 57 of , following the signal peptide, could not be modelled in the absence of suitable template for that part of the sequence. The best among them were chosen depending on the DOPE score, ERRAT score, PDBSum analysis and ProSA-web protein structure analysis (Table 1). The models having best validation parameters chosen for further study are shown in figure 1. NDUFS2 and 7 are primarily helical structures with a few β-sheets, while NDUFS3 and 8 comprise of loops and β-sheets, along with few helices. |
| The mutant structures for all the four subunits of Q module were generated using in silico mutagenesis. The RMSD were insignificant for residues forming secondary structures in the post energy minimized models of these mutants. |
| Molecular dynamics simulation of subunit and mutant structures |
| The effects of various reported mutations were evaluated by using MD simulations over a 15 ns trajectory. The RMSD corresponding to the Cα atom positions of each mutant, with respect to the wild-type subunit, were compared (Supplementary Figure SF5 a-d). Any changes in the hydrogen bonded interactions on mutating each subunit, followed by energy minimization, were identified. |
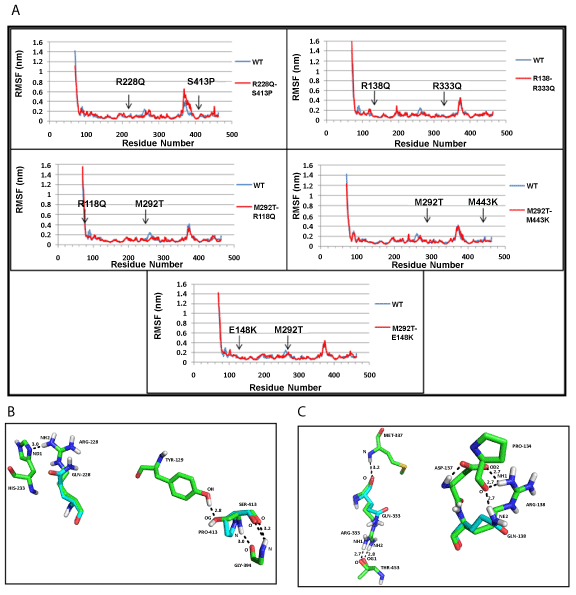
| NDUFS2: The root mean square fluctuations (RMSF) indicated minor changes in the positions of amino acid residues present at the mutation sites (Figure 2a). In one of the mutants (R228Q+S413P), the hydrogen bond formed between amino acid residues, R228 and H223, was lost with R228Q mutation; similarly, the hydrogen bonded interaction between S413 and Y129 was lost by S413P mutation (Figure 2b), in mutant (R138Q+R333Q), the first mutation destroyed side chain ionic interaction between amino acids R138 and D137 (Figure 2c). No change in hydrogen bonds were found in the remaining three mutants: (M292T+E118Q), (M292T+M443K), (M292T+E148K). The overall compactness of the structure, as indicated by the Radius of gyration (Rg) (Supplementary figure SF6a), remains fairly constant at the end of the 15 ns simulation for the wild-type protein and mutants. |
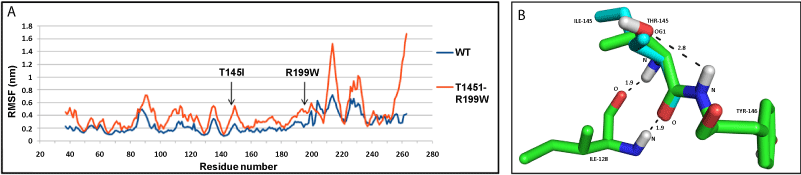
| NDUFS3: The values of RMSF indicated that the mutant (T145I+R199W) structure had a higher number of fluctuating residues, as compared to the wild-type (WT) protein (Figure 3a). This may be accounted for by the large number of loops present in NDUFS3 structure. At the end of the simulation, WT protein shows a more compact structure, compared to the mutant as measured by the radius of gyration (Rg) (Supplementary Figure SF6b). The hydrogen bond between amino acid residue, T145 and Y146, was lost by the mutation T145I; however, no changes were observed for R199W mutation (Figure 3b). |
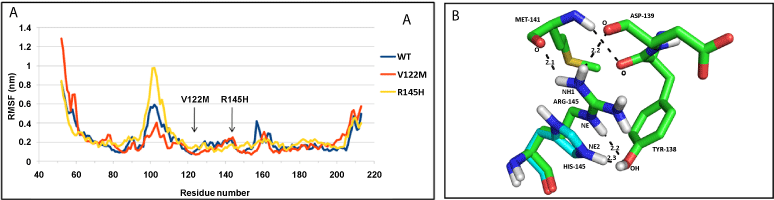
| NDUFS7: Both the mutant structures (V122M and R145H), show difference in fluctuation of residues, compared to WT protein throughout the structure (Figure 4a). The WT protein and mutants have similar compactness measured by Rg (Supplementary figure SF 6c). V122M mutation did not influence hydrogen bonds, whereas the R145H mutation disrupted hydrogen bonds of R residue with M141 and D139 (Figure 4b). |
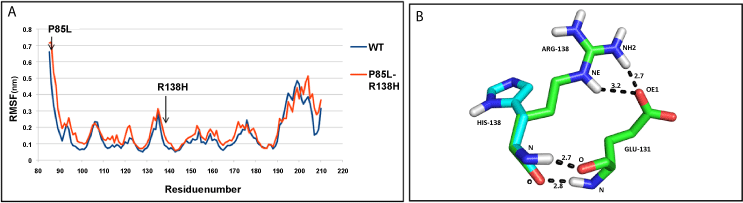
| NDUFS8: The residue-fluctuations were more in mutant (P85L+R138H), compared to the WT protein across the structure (Figure 5a). At the end of simulation, the values of Rg pointed to a more compact WT structure than mutant (Supplementary figure SF 6d ). The hydrogen bond between residues R138 and E131 was lost, as a result of R138H mutation (Figure 5b). |
| The changes in the solvent accessible area, as well as average intraand inter-molecular hydrogen bonds, are listed in table 2. In all the mutants of NDUFS2 and , a distinct increase or decrease in the solvent accessible area of mutant compared to WT was seen. Although there was no change in solvent accessible area of mutant compared to WT of NDUFS3, WT was more compact than mutant. Similarly, in the case of NDUFS7, the R145H mutant did not show any change in the solvent accessibility. |
| The change in the intra- and inter-molecular hydrogen bonds of WT and mutants during the 15 ns simulation, and their average values at the end of the trajectory are listed in table 2. In each case, different changes were observed in either the number of intra- or inter-molecular hydrogen bonds formed by WT compared to mutants. The changes in the number and nature of hydrogen bonds could be considered an indication of the changes to atomic positions in WT and mutant structures taking place through the 15 ns simulation. On examining the time evolution of structures during the 15 ns trajectory, no major changes in the secondary structure between mutants and WT could be detected. |
| Molecular model of Q module assembly |
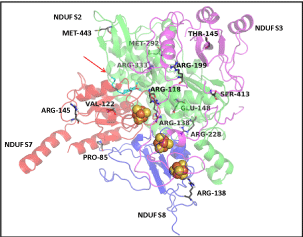
| The conserved residues at the interface of four subunits in the Q module assembly modelled (Figure 6) were examined. Most interacting interface residues of the Thermus thermophilus are found conserved in the human mitochondrial Q module, as well. The three iron-sulphur clusters modelled within the Q module remains the path for the entry of DBQ, indicating importance of the correct assembly in complex formation. |
| Binding of n-decyl-ubiquinone (DBQ) |
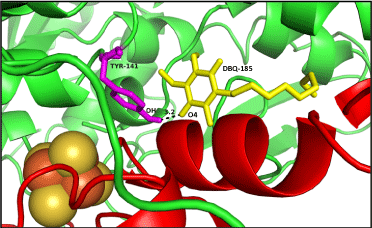
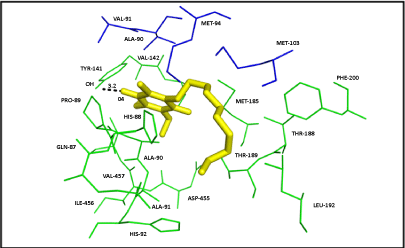
| Ten poses were generated in Glide5.8 for DBQ docking in Q module. The glide gscore and emodel score (estimated conformational energy of the ligand) for the best pose (Figure 7) was -6.08 and -31.07, respectively. The ligand interaction diagram for DBQ is a two-dimensional plot, showing the interactions of the ligand bound to the protein. The residue Y141 (equivalent to Y144 of yeast and Y87 of Thermus), close to the iron-sulphur cluster N2, forms one hydrogen bond with the carbonyl head group of DBQ, and the hydrophobic side chain interacts with the hydrophobic residues at the interface of NDUFS2 and NDUFS7 (Figure 8). Amino acid residues T189, L192, F200 and V457 of NDUFS2, and M94 and V91 of NDUFS7, are in the proximity of DBQ. It is interesting to note that site directed mutagenesis study in yeast have shown the importance of these residues in binding DBQ [7]. |
| Residue scanning of Q module complex and energy calculations |
| The differences in energies between the WT and mutant complexes of Q module were calculated in Bioluminate 1.0 residue scanning module. The contribution of individual mutations toward the energy and stability of the complex was estimated, in terms of differences in energies of mutant and WT protein structures (Table 3). The Δ stability calculations measures the instability of mutant structures compared to WT. All mutations except R118Q, showed positive values of Δ stability, indicating the role mutations play as structure destabilizing factor. The NDUFS2 mutation S413P could not be tested because Bioluminate program had no provision to mutate from non-proline residue to proline. Estimate of the binding affinities of mutants towards iron-sulphur clusters, DBQ and neighbouring subunits indicated a reduction in affinity of certain mutants, such as E148K of NDUFS2 and P85L of towards iron-sulphur clusters. Similarly, a reduction in affinity towards DBQ was observed for most mutations of NDUFS7 and mutations of NDUFS2, except R138Q, E148K and M443K. Majority of the mutations were affecting the inter-subunit affinity and stability of the complex (Table 3). |
| Discussion |
| Complex-I is known as a hot spot for mutations, leading to several mitochondrial diseases. The list of reported mutations is ever increasing in human mitochondrial genome database: Mitomap [35]. Proper assembly and stabilization of Complex-I is essential for its activity, as well as structural stability of the inner mitochondrial membrane. Deleterious mutations perturb the assembly of Complex-I and induce structural changes in the mitochondrial membrane, causing proton leakage, accumulation of reactive oxygen species, and thereby, release of apoptotic factors which remain primary causes of these diseases [36]. Reports of BN-PAGE studies indicate that the mutations affect the assembly process at an early stage, resulting in the accumulation of assembly intermediates. The stages of subunit association are determined, such that NDUFS2 and NDUFS3 assemble first, followed by NDUFS7, and finally , which indicates that a mutation in any one subunit would affect assembling into full functional module [37]. |
| We classified the known mutations into four categories: as mutations of the internal residues, mutations of the surface residues, mutations at the subunit interface and mutations close to binding sites (Table 4). NDUFS2 and NDUFS7 act as functional subunits of the Q module, whereas the role of NDUFS3 and are more of a structural nature. Hence, during the MD simulation, drastic structural changes were observed in the NDUFS3 and 8, whereas NDUFS2 and 7 showed instability contributing from energy changes associated with function. |
| The trajectory of 15 ns simulation revealed several structural changes caused by mutations. The structures of NDUFS3 and displayed an overall increase in fluctuation of residues in their mutant structures, probably owing to the large number of loops present in these structures. Also the structures of the WT protein of both subunits remained more compact, compared to the corresponding mutants. In NDUFS7, the residue fluctuations at the points of mutation were accompanied with large fluctuations in the loop regions, whereas NDUFS2 had fluctuations only at the points of mutations. Thus, the influence of the mutations on the flexibility of polypeptide chain was confined to the points of mutations in NDUFS2, but extended to neighbourhood in others. Interestingly, mutations such as M292T+R118Q, M292T+M443K and M292T+E148K of NDUFS2 and V122M of NDUFS7 rarely affected hydrogen bonds at the points of mutations, although changes were reflected in the average number of intra- or inter-molecular hydrogen bonds (Table 2). The interior residue mutation M292T is found in combination with different other mutations of NDUFS2, in four unrelated individuals of Caucasian families suffering from Leigh syndrome [9]. This mutation did not affect hydrogen bond with neighbourhood residue G290. However, it introduced an additional protein kinase C phosphorylation site at threonine, potentially affecting the regulatory function of the Q module [38]. Two NDUFS2 mutations, interior residue E148K and surface residue M443K, although did not influence the hydrogen bonds, affected the affinity for the iron-sulphur clusters (Table 3). |
| The T145I interior residue mutation in NDUFS3 was at a putative casein II phosphorylation site [38], and caused loss of a critical hydrogen bond, thereby decreasing the protein stability. The surface mutation R199W, occurring in combination with interior residue mutation T145I, decreased the affinity of the NDUFS3 for other subunits, thereby decreasing the overall stability of the Q module. The interesting observation is that the patient inherited individual mutations from each healthy parent, thereby showing that the combination only caused disease [11]. Analysis of the post simulation structures indicated that the T145I mutation resulted in a shift in the secondary structure. When it occurred as a combination with the other mutation R199W, it resulted in the shifting of the loop, containing amino acid residues 196-206 towards the NDUFS2 subunit, causing a major deviation of the loops and β-sheets along the structure. Severe steric clashes with subunits, NDUFS2 and of assembly, were also observed. Thus, when T145I mutation occurs together with R199W, the NDUFS3 subunit becomes highly unstable, evidenced by the increased fluctuations of residues and decreased compactness. The loops turning highly dynamic affect interaction with NDUFS2 subunit, preventing the initiation of assembly process. |
| The mutation V122M in the functionally important subunit NDUFS7 had no effect on any hydrogen bond. The mutated residue lies within a 4 Å radius of the iron-sulphur cluster N2, located within the cysteine motif-CxxE-(X)60-C-(X)30-CP-, and involved in the formation of N2 cluster [12]. Irrespective of its proximity, the mutation did not change affinity for the iron -sulphur clusters. Surprisingly, the mutation slightly decreased the affinity for DBQ (Table 3), although not lying in the immediate vicinity of DBQ binding site, indicating the importance of this residue for the function of subunit. The surface mutation R145H not only caused loss of a hydrogen bond, but also reduced the affinity towards DBQ. Thus, the point mutations in NDUFS7 severely affect the activity of Complex-I by reducing affinity towards DBQ. |
| The interface mutation, P85L of , reduced the affinity of the module towards iron-sulphur clusters. Similarly, the other surface mutation R138H resulted in removal of hydrogen bond with residue E131, thereby decreasing the stability of subunit. The patient bearing these two mutations was homozygous for both mutations, which had been inherited one each from healthy heterozygous parents, indicating lethality of their combination to cause disease [14]. |
| The residues R118, R138, R228 of NDUFS2, and P85 and R138 of , were present at the interface of the 4 subunits (Table 4). In accordance with their positions, the mutations will have deleterious effect on the inter-subunit affinity, as shown by the energy calculations in Bioluminate 1.0, except for mutants (Table 3). The positions of the mutations, with respect to the iron-sulphur clusters and docked DBQ, indicated that amino acid residues R118, R138, R228 of NDUFS2, V122 of NDUFS7 and R138 of , were fairly close to ligands (Figure 6). The energy calculations in Bioluminate 1.0 reflect destabilizing effect of these mutations. |
| Large changes in RMSF, Rg and observed differences in the hydrogen bond formation showed the lethal effect of these point mutations on the structure of the individual subunits. Thus, the mutant molecule, with initial secondary structure similar to wild-type protein, tends to be distinctly different during MD simulation. The effect of these point mutations on the assembly and function of the Q module studied gave further insight on the consequences of these mutations on the structure assembly, as a whole. However, it would be interesting to note that some of these mutations, especially in those residues is structurally interior, need to occur in combination to show up as disease phenotype, confirming that their cumulative effect what is causing disease condition. The stability values also show that most of these mutants are energetically highly unstable. In NDUFS3, and NDUFS8, some single mutations alone were not lethal, as they were present in healthy parents, reflected in the Δ affinity values that were not significant (Table 3). Hence, their effects are more pronounced only when in combination. In silico structural and functional analysis of the Q module showed that the mutations affected the system structurally by the mutated region, tending to become more flexible, changing the compactness of the structure and resulting in loss of some hydrogen bonded interactions, accompanied by an increase in energy; the mutations functionally affect the system by reducing affinities towards the iron-sulphur clusters, DBQ and with the other core subunits. The accumulation of the assembly intermediates observed in patients having such mutations is the net effect of all the factors described here, with respect to the structure and function of the Q module. |
| Acknowledgements |
| Tulika M Jaokar and Ranu Sharma thank Council of Scientific and Industrial Research (CSIR), India, for research fellowship. Funding from CoESC programme of CSIR-NCL is acknowledged. |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 | Table 4 |
Figures at a glance
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
 |
 |
 |
 |
| Figure 5 | Figure 6 | Figure 7 | Figure 8 |
Relevant Topics
- Analytical Biochemistry
- Applied Biochemistry
- Carbohydrate Biochemistry
- Cellular Biochemistry
- Clinical_Biochemistry
- Comparative Biochemistry
- Environmental Biochemistry
- Forensic Biochemistry
- Lipid Biochemistry
- Medical_Biochemistry
- Metabolomics
- Nutritional Biochemistry
- Pesticide Biochemistry
- Process Biochemistry
- Protein_Biochemistry
- Single-Cell Biochemistry
- Soil_Biochemistry
Recommended Journals
- Biosensor Journals
- Cellular Biology Journal
- Journal of Biochemistry and Microbial Toxicology
- Journal of Biochemistry and Cell Biology
- Journal of Biological and Medical Sciences
- Journal of Cell Biology & Immunology
- Journal of Cellular and Molecular Pharmacology
- Journal of Chemical Biology & Therapeutics
- Journal of Phytochemicistry And Biochemistry
Article Tools
Article Usage
- Total views: 13736
- [From(publication date):
specialissue-2013 - Apr 09, 2025] - Breakdown by view type
- HTML page views : 9132
- PDF downloads : 4604
