Research Article Open Access
Spatial Analysis of Greater Sage-grouse Habitat Use in Relation to Landscape Level Habitat Structure
Mark T Freese1, Steven L Petersen2*, Richard F Miller3, Andrew C Yost4 and W Douglas Robinson51Wildlife Biologist, Nevada Department of Wildlife, Valley Road, Reno, Nevada, USA.
2Department of Plant and Wildlife Sciences, Brigham Young University, WIDB, Provo Utah, USA.
3Department of Animal and Rangeland Sciences, Oregon State University, Withy combe Hall, Corvallis Oregon, USA.
4Oregon Department of Forestry, State Street, Salem, USA.
5Department of Fisheries and Wildlife, Oregon State University, Nash Hall, Corvallis, USA.
- *Corresponding Author:
- Steven Petersen
Department of Plant and Wildlife Sciences
Brigham Young University, 487 WIDB
Provo Utah 84602, USA
Tel: (801) 422-4885
E-mail: steven_petersen@byu.edu
Received Date: July 05, 2016; Accepted Date: July 29, 2016; Published Date: August 05, 2016
Citation:Freese MT, Petersen SL, Miller RF, Yost AC, Robinson WD (2016) Spatial Analysis of Greater Sage-grouse Habitat Use in Relation to Landscape Level Habitat Structure. J Ecosys Ecograph 6:205. doi:10.4172/2157-7625.1000205
Copyright: © 2016 Freese MT, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Ecosystem & Ecography
Abstract
Greater sage-grouse (GSG; Centrocercus urophasianus) selectively utilize portions of sagebrush and sagebrush associated habitats within broad and heterogeneous landscapes. Until recently, sage-grouse research has generally focused on fine-scale vegetation structure and composition and less on landscape-scale habitat requirements. Insufficient information at broad scales limits a manager’s ability to interpret and predict habitat use patterns, assess habitat suitability, and target areas for conservation and ecological rehabilitation. We identified environmental attributes associated with GSG habitat use at broad spatial scales. In 2006, we captured 50 GSG, radio-collared each bird, and tracked each bird’s position within a 31,416 ha study area in central Oregon, USA. We monitored birds year-long between March 2006 and March 2008 across the study area. Each time a bird was located, we collected a coordinate position at the point where it was observed. We generated spatially explicit predictor variables in a Geographic Information System to quantify the association between landscape structure and GSG occurrence. Predictor variables included elevation, slope, aspect, curvature, solar radiation, landscape ruggedness, orientation, distance from roads, distance from leks, distance from mesic habitats, and cover type. We used spatial modeling (Maximum Entropy) to 1) develop predictive models of GSG seasonal resource use, 2) generate probability maps for visual assessment, and 3) characterize response curves associated with GSG habitat preference based on individual landscape predictor variables. Results indicate that during the breeding season GSG will use big sagebrush, low sagebrush or complexes of low and mountain big sagebrush cover types. During the summer season, GSG use low sagebrush, mountain big sagebrush, and mesic areas. Additionally, summer season use areas include higher elevation sites within or in close proximity to habitats that sustain succulent forbs throughout most of the growing season. Maps of modeled data identify spatially explicit areas of preferred habitat and predicted bird use patterns. This information can help managers identify and protect important GSG habitat across heterogeneous landscapes.
Keywords
Centrocercus urophasianus ; Greater sage-grouse; Habitat; Landscape-scale; Modeling; Radiotelemetry
Disclaimer
The findings and conclusions of this article are those of the author(s) and do not necessarily represent the views of the U.S. Fish and Wildlife Service.
Introduction
Greater sage-grouse (GSG; Centrocercus urophasianus ) habitat research has focused primarily on fine-scale (0.007 - 0.032 ha) vegetation structure and composition immediately surrounding sites where birds have been observed. This research has been instrumental in explaining the vegetative components necessary for GSG survival and reproduction [1-5]. However, GSG utilize broad landscapes where land management agencies have responsibility of making decisions and planning conservation strategies that are relevant at the landscapelevel [6-7]. There is often insufficient information about GSG habitat requirements that can be accounted for at the landscape-level, limiting a manager’s ability to interpret and predict habitat use patterns, assess habitat suitability in meaningful locations (i.e., versus random locations), and target areas for conservation and ecological restoration across broad heterogeneous landscapes.
Greater sage-grouse select habitat at multiple spatial scales with home ranges that can exceed 270,000 ha [8-10]. The ability of GSG to move long distances allows them to utilize the spatial and temporal variation in sagebrush and associated plant community resources [11]. This movement is often a display of complex selection patterns that result from variation in the composition, structure, and arrangement of sagebrush ecosystems. This heterogeneity in sagebrush communities can create a challenge for managers as they identify manageable features of GSG habitat that attract greater bird use and provide a consistent supply of resources that meet the nutritional and safety requirements of a sustainable population.
Similar to GSG, greater prairie chickens (Tympanuchus pallidicinctus ) display characteristic movement and habitat use patterns throughout the year as nesting activity transitions to brood rearing. According to Fuhlendorf et al. [12], there is no single spatial scale sufficient to characterize the influences of landscape structure on greater prairie chicken habitat use. Instead, they emphasize the need for research and management that covers multiple spatiotemporal scales. Since GSG utilize resources across expansive and heterogeneous landscapes, methods are needed that accurately identify the attributes most closely associated with GSG habitat use patterns. Previous research [13] has shown that GSG habitat can be modeled as a function of landscape structure in an ecological niche modeling framework using Maxent species distribution modeling software. Ecological niche models (ENM) are a class of methods that use species occurrence data and associated environmental conditions to 1) estimate the relative suitability of habitat known to be occupied by the species, 2) estimate the relative suitability of habitat in geographic areas not known to be occupied by the species, 3) to estimate changes in the suitability of habitat over time, and 4) estimate the actual niche of a species [14]. The purpose of this research is to quantitatively model and map breeding areas and suitable brood rearing habitat in a sagebrush- (Artemisia tridentate Nutt. ) dominated plant community of central Oregon using a set of landscape-level attributes, GSG telemetry locations, and Maxent species distribution modeling software.
Methods
Study area
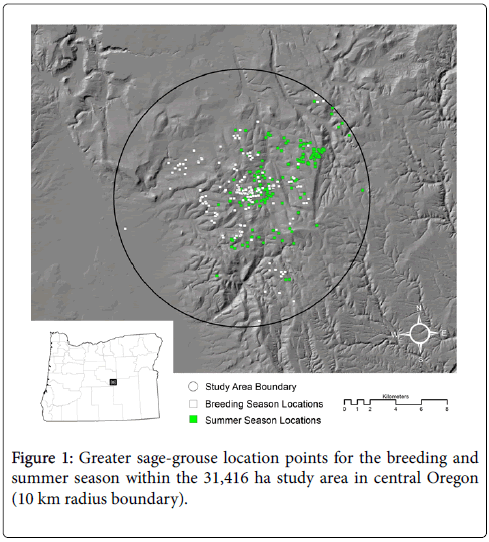
The study area is located in Crook County of central Oregon (Figure 1) and supports a population of GSG that have been relatively stable since 1987. Plant communities are characterized by Wyoming big sagebrush (A. tridentata ssp. wyomingensis Beetle and Young) at lower elevations and a complex of low sagebrush (A. arbuscula Nutt.) and mountain big sagebrush (A. tridentata spp. Vaseyana (Rydb.) Beetle) at higher elevations. Wet and dry meadows and riparian communities occur in scattered patches throughout the region, accounting for <1% of the landscape.
In the center of the study area is the largest of three adjoining leks (119º53’ 22” W 43º48’ 31” N). During past breeding seasons (1994-2008), this lek was visited by an average of 47 males (range 27-71). The spatial extent of the study area is represented by a circle with a 10 km radius typical of post-lekking dispersal distances described by Braun et al. [15] and Connelly et al. [8]. Similarly, Aldridge and Boyce [16] found that 90% of all nesting and broodrearing habitats occurred within 10 km of a lek and Bruce et al. [17] found that 85% of the collared birds remained within 10 km of the lek during the breeding and summer seasons at this same study area.
The study area is composed of both private (55%) and public (45%) lands. The public land is divided into lands managed by the Bureau of Land Management (35%), the United States Forest Service (6%), and the state of Oregon (4%). The geology and soils resemble the High Desert and John Day Ecological provinces [18]. The topography is characterized by elongated ridges with dissecting draws, rolling hills, rocky tablelands, and interspersed with buttes and plateaus capped with basalt or tuffaceous rock. Dominant soils are comprised of mesic and frigid temperature and xeric moisture regimes that include Argixerolls, Haploxerolls, Palexerolls, Haploxerents, and Durixerolls. Elevation generally increases from west to east ranging from 1,267 m to 1,715 m. Annual precipitation varies between 20-40 cm. Average annual temperature is 7.5°C (range −29 to 38°C) and precipitation for the water year (October 1 – September 30, 2007) was 59% of the longterm average (EOARC weather station records 1971-2000).
Greater sage-grouse location data
We captured adult and subadult GSG from March 2006 through March 2007 during the lekking period. Birds were opportunistically located during nighttime hours using a spotlight powered by a Honda 350x backback generator. Once netted, each bird was gently handled and fitted for a VHF collar as quickly and carefully as possible to minimize stress. Each bird was fitted with a 22 g self-releasing collar. Additional description of trapping and collaring methods are described by Giesen et al. [19]. We trapped 30 male and 20 female birds at six leks or roosting areas. Each bird was fitted with an Advanced Telemetry Systems (ATS) radio transmitter for monitoring its location after release using a portable antenna/receiver and GPS. Each collared bird was located on average every 15 days (range 1-154 days). We recorded 250 location points during three breeding seasons from March through June in 2006, February through June in 2007, and February through March in 2008. We recorded 207 location points during two early and late brood-rearing seasons from June through October in 2006, and June through October in 2007). These seasons represent temporal categories used to distinguish variability in the amount and quality of habitat and resources (i.e., insect, forb and water availability) required by GSG [8,20,21].
Analysis
Predictive Modeling with Maxent — We used maximum Entropy (Maxent) to generate seasonal habitat suitability models and maps using GSG locations and a set of landscape-level predictor variables. For input MaxEnt uses a list of presence locations of a particular species in combination with a set of environmental predictor variables across a user-defined landscape represented as an array of grid cells. From this landscape, predictor variable information is extracted from a spatially diffuse sample of background locations, where presence is unknown, and is contrasted against the predictor variable information at presence locations. Thus, MaxEnt predicts the relative occurrence rate of a species for any one grid cell as a function of the environmental predictors. MaxEnt derives mathematical transformations of each predictor (linear, quadratic, product, hinge, threshold), referred to as features, that users can select from to generate complex, nonlinear models using all features or simpler models with fewer features. Use of the full set of feature types can lead to highly complex models and overfitting depending on the sample size of the training data. For example, use of the threshold feature requires a minimum of 80 training samples and can produce sharp jumps in response curves that manifest in maps with odd discontinuities [22]. Categorical predictors with n categories are split into n binary features, which take the value 1 when the feature is present and 0 otherwise.
To obtain a solution, MaxEnt maximizes a penalized maximum likelihood function referred to as the gain. The exponentiated gain function represents the likelihood ratio of an average presence to an average background point. Maximizing the gain, therefore, corresponds to finding a model that best differentiates presences from background locations that are uniformly distributed across the study area. The gain represents the sum of predicted values at presence locations from which the log of the sum of predicted values at background locations and an overfitting penalty are subtracted [23]. Gain is a measure of likelihood analogous to deviance in Generalized Linear Models and can be interpreted as the degree of improvement in the Maxent probability distribution fitting the sample points compared to a uniform distribution. For example, if the gain is 1, the average sample likelihood is exp (1) = 2.7 times higher than that of a random pixel. For the full mathematical description of the MaxEnt algorithm readers should see Phillips et al. [24], Merow et al. [23], or Elith et al. [25].
Predictor variables
A 10 m Digital Elevation Model (DEM) was used to generate raster layers for the predictor variables elevation, ruggedness index, aspect, slope, curvature, and hillshade, with the latter four of these generated using the Surface tools available in the Spatial Analyst Toolbox of ArcGIS®. The ruggedness index represents a measure of terrain ruggedness derived from the variation in three-dimensional orientation of grid cells within a specified neighborhood. This index captures variability in slope and aspect into a single measure. Ruggedness values in the output raster can range from 0 (no terrain variation) to 1 (complete terrain variation). The ranges of terrain values for the study area were typical values for natural terrains between 0 and about 0.4 [26]. Aspect values were categorized as flat, north (316° to 45°), east (46° to 135°), south (136° to 225°), and west (226° to 315°). The curvature raster represents the convexity and concavity of each cell based on the mean elevation of a 5 × 5 window centered on the target cell and was created with the “curvature” function. A positive curvature indicates the surface is upwardly convex at that cell. A negative curvature indicates the surface is upwardly concave at that cell. A value of 0 indicates the surface is flat. The slope raster was derived using the “slope” function. The “hillshade” function was used to generate the hypothetical solar illumination on each cell of the raster. We compared the training gain produced with six values for the highest altitude of the sun with the corresponding azimuth associated with the first day of each month from January through July but chose the default settings provided in the ArcGIS®. Hillshade tool because it produced the highest training gain for both the breeding and summer season datasets.
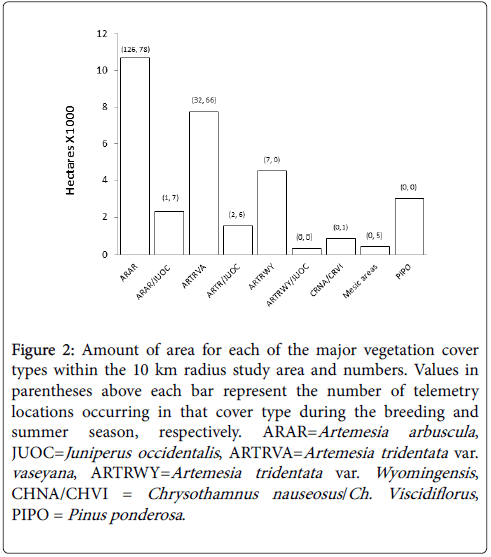
Vegetation cover types were delineated from 0.5 m National Agriculture Imagery Program (NAIP) aerial color photographs and attributed with data collected from field-based observations. GIS polygons of each cover type were digitized within a 10 km radius (314 km2) centered at the site of highest courtship activity (Figure 1). The accuracy assessment conducted to compare the actual vegetation type to the predicted type from 40 randomly selected GIS locations produced a “better than substantial” Kappa statistic of 0.89 [27]. This analysis resulted in a GIS layer with 18 different field-sampled cover types. For the purposes of generating Maxent models we wanted to avoid the potential inflation of prediction values, therefore, cover types comprising less than 1.5% of the study area were merged with the most similar type (Figure 2). This integration resulted in a categorical predictor variable with nine major vegetation cover types.
Figure 2: Amount of area for each of the major vegetation cover types within the 10 km radius study area and numbers. Values in parentheses above each bar represent the number of telemetry locations occurring in that cover type during the breeding and summer season, respectively. ARAR=Artemesia arbuscula , JUOC=Juniperus occidentalis , ARTRVA=Artemesia tridentata var. vaseyana , ARTRWY=Artemesia tridentata var. Wyomingensis , CHNA/CHVI = Chrysothamnus nauseosus /Ch. Viscidiflorus , PIPO = Pinus ponderosa
A predictor variable representing the distance from roads was generated by calculating the Euclidean distance of each raster cell from digitized road features and another representing distance from streams which include locations attributed as perennial streams, wet meadows, and stock-pond/reservoirs.
Two variables representing east and north UTM coordinates were created to compare the performance of models built with and without location coordinates. While both types of models are useful for predicting where a species might occur in unsampled areas, models with location coordinates are useful for generating maps that emphasize the spatial distribution of the presence data. Models without coordinates are more useful for generating habitat suitability maps to help explain why a species occurs where it does and how it responds to the ecological and geophysical gradients represented with the predictor variables.
Model Development
The overall modeling objective was to build habitat suitability models with high performance from the best subset of predictor variables chosen for this analysis. We first generated a set of models that included the north and east UTM predictor variables for depicting the strongest spatial distribution of locations and evaluating the extent of spatial autocorrelation. We generated another set of models without spatial coordinates to identify which of the remaining variables were most important in predicting suitable habitat.
Maxent’s machine learning approach of including all reasonable predictors in a model and letting the algorithm decide which ones are important via regularization can result in highly complex models even with prescreening of correlated variables. Therefore, to produce simpler models that have similar performance to more complex models [23] we followed the method for selecting the best subset of predictor variables used by Yost et al. [13]. The technique was found to be simple, yet effective, to objectively determine which variables made insignificant contributions to a model. Maxent's jackknife test of variable importance can be used to evaluate the relative strengths of each predictor variable. The training gain is calculated for each variable alone and the drop in training gain when the variable is omitted from the full model. The modeling process started with a full model containing all predictor variables. Then, the variable with the lowest decrease in the average training gain when omitted from the full model was eliminated, and the remaining variables were used to build a reduced model. This process was repeated until the average training gain for the reduced model dropped significantly below the average training gain for the full model or the model with the highest training gain. Overlap between 95% confidence intervals for training gain averages was the criteria for significance. Averages and confidence intervals were estimated from 10 bootstrapped random replicates with 75% of the presence records used for model calibration and the remaining 25% for testing.
Notwithstanding drawbacks reported by Lobo et al. [28] we used Maxent’s calculation of the area under the receiver operating curve (AUC) to report model accuracy and fit [29]. The AUC is an index representing the model’s predictive accuracy that ranges in value from 0.5 to one. Models with AUC values >0.9 can be considered to have excellent predictive ability, between 0.8 and 0.9 as good, 0.7 to 0.8 as fair, 0.6 to 0.7 as poor and 0.5 to 0.6 represents failure or models that don’t predict better than a random guess [30]. Model success was also evaluated by visually inspecting how well the probability values in the output grid fit with GSG location points. A good model will produce regions of high probability that cover the majority of presence records and areas of low probability should contain few to no presence points.
We used linear, product, and hinge features and selected the auto features option. Default values were used for the Maxent algorithm to approach convergence based on the maximum number of iterations (1000), the convergence threshold of 10-5, the regularization multiplier to 1.0, and prevalence to 0.5. Data were pooled across years and gender for each season and multiple observations of birds at the same location were treated as one data point (by selecting the Maxent option to remove duplicate presence records) used for model building resulting in a total of n = 168 (17 female, 26 male) for the breeding season and n = 163 (16 female, 21 male) for the summer season. The number of observations of individual birds throughout the study ranged from 1 to 17 with an average of 5.5 (± 1.3).
We used the lowest average prediction value from the Maxent output that corresponded with the area adjusted frequency [31] significantly greater than one to objectively reclassify the continuous logistic output into a binary map of “suitable” and “unsuitable” habitat classes [22]. The area-adjusted frequency, hereafter referred to as the predicted to expected (P/E) ratio [32], was estimated by first partitioning the range of corresponding logistic values from Maxent’s omission output file into 91 classes (0.05 incremented by 0.01 to 0.95). The prediction value for each class was estimated with linear interpolation between a range of corresponding logistic values defined by a width of W = 0.1 [31]. Computation started with a first class covering the class values that ranged from [0,W]. The numerator of the P/E ratio was the interpolation between the test omission value antecedent to the one that corresponded with the lower boundary of the computation width and the one that corresponded with the upper boundary. The denominator was the interpolation between the corresponding fractional area values. The P/E ratio was calculated for each subsequent class that expanded with each increment up to a width size of [W/2 – class value + W/2] and ended with a size of [1-W, 1]. The P/E ratios were then averaged across all replicate models, and 95% confidence intervals calculated. We considered the class subsequent to the last class with a lower confidence limit that overlapped the P/E = 1 as significant and used it as the map reclassification value. If the habitat model properly delineates the species suitable areas, a low suitability class should contain fewer evaluation presences than expected by chance, resulting in P/E <1. Conversely, high suitability classes should have P/E increasingly higher than 1 [31,32].
Ethics statement
This study was carried out with strict accordance with the protocol approved by the Oregon State University Institutional Animal Care and Use Committee. A sage-grouse trapping/handling permit was obtained from the Oregon Department of Fish and Game prior to trapping efforts. Plant data was collected with permission provided by the land owner and manager. The research did not involve federally or state listed endangered species.
Results
Breeding season model
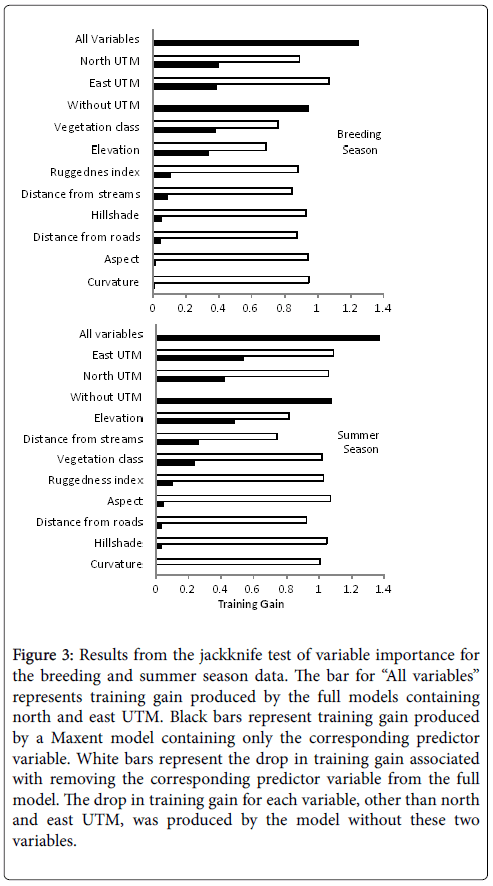
We removed the predictor variable for slope from the modeling process because the correlation with the ruggedness index was greater than 0.7 when included and it reduced the training gain less when removed from the model. Similarly, elevation was also removed because it was correlated (0.7) with East UTM and reduced the training gain less when removed from the full model (Figure 3). The average AUC value for the nine-variable model was 0.904 with an average training gain of 1.25. The Jacknife test of variable importance shows that the individual training gains for north UTM (0.403), vegetation classification (0.4) and east UTM (0.39) were nearly the same value and highest among the full set of predictor variables. The UTM coordinates decreased the training gain the most when omitted from the full model followed by the vegetation classification (Figure 3).
Figure 3: Results from the jackknife test of variable importance for the breeding and summer season data. The bar for ��?All variables� represents training gain produced by the full models containing north and east UTM. Black bars represent training gain produced by a Maxent model containing only the corresponding predictor variable. White bars represent the drop in training gain associated with removing the corresponding predictor variable from the full model. The drop in training gain for each variable, other than north and east UTM, was produced by the model without these two variables.
To simplify the full model and reduce the possibility of overfitting we removed variables one at a time based on the least amount of decrease in the average training gain when a particular variable was omitted. There was overlap in 95% confidence intervals even though the average training gain (in parentheses) varied through the removal of curvature (1.25), aspect (1.34), hillshade (1.24), and distance from roads (1.24). Training gain dropped (1.09) significantly and eliminated the overlap in confidence intervals with the full model when ruggedness index was removed. Therefore, the final model contained the UTM coordinates, vegetation classification, distance from streams, and the ruggedness index (Table 1).
| Breeding Season | Summer Season | |||
|---|---|---|---|---|
| Model 1 | Model 2 | Model 1 | Model 2 | |
| Training AUC | 0.90 | 0.85 | 0.91 | 0.88 |
| Test AUC | 0.87 | 0.84 | 0.88 | 0.86 |
| Training Gain | 1.24 | 0.85 | 1.25 | 1.0 |
| Test Gain | 1.24 | 0.96 | 1.20 | 0.97 |
| PredictorVariables | North UTM East UTM Vegetation Dist. Streams Ruggedness |
Vegetation Elevation Ruggedness Dist.Streams |
North UTM East UTM Dist. Streams Dist. Roads |
Elevation Dist. Streams Vegetation Ruggedness Dist. Roads |
Table 1: Evaluation statistics and predictor variables comprising models with UTM coordinates (Model 1) and those without coordinates (Model 2) for the breeding and summer season datasets.
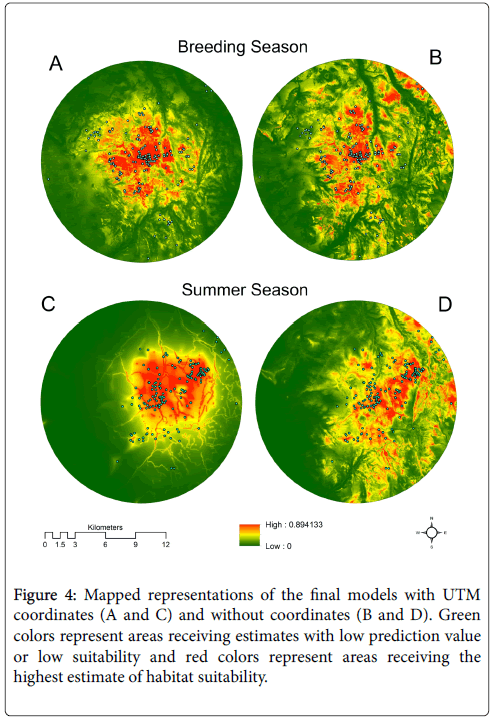
The final model had excellent predictive ability with an average AUC of 0.90, predicted areas used for breeding about 3.5 times better than a random distribution, and was used to generate the distribution map in Figure 4A. The reclassification procedure resulted in designating prediction values greater than 0.41 (9.1% of the study area) as suitable habitat and prediction values less than or equal to 0.41 as unsuitable habitat.
Breeding season without UTM coordinates
The AUC for the full model without the UTM predictor variables was 0.874 and the average training gain was 0.95. The predictor variable for elevation was included in the full model because the correlation with East UTM was no longer a factor. The jackknife test of variable importance showed that the vegetation classification variable had the highest average training gain (0.39) when used alone followed by elevation (0.35). Conversely, elevation produced the largest drop in training gain when removed from the full model followed by vegetation. Training gain values among the other predictors were considerably lower not exceeding 0.11.
There was overlap in the confidence intervals for the average training gain with the full model through the removal of curvature (0.96), aspect (0.99), hillshade (0.93), and distance from roads (0.85) but with the removal of the ruggedness index (0.71) the drop in average training gain was significant eliminating the overlap in confidence intervals. The final model contained vegetation, elevation, the ruggedness index, and distance from streams (Table 1). The AUC value for the final model was 0.851. It predicted breeding habitat about 2.3 times better than a random distribution, and was used to generate the map of breeding habitat (Figure 4B). The reclassification procedure identified prediction values greater than 0.52 (9.1% of the study area) as suitable habitat.
Summer season
The AUC for the full model without was 0.92 and the average training gain was 1.38. The predictor variable for elevation was not included in the full model because the correlation with East UTM. The Jacknife test of variable importance shows that the individual training gains for north UTM (0.43) and east UTM (0.54) were again the highest among the set of predictor variables and lowered the average training gain the most when removed from the full model. The next highest training gain was associated with distance from streams (0.30) followed by the vegetation classification (0.25).
There was overlap in the confidence intervals for the average training gain with the full model through the removal of aspect (1.37), curvature (1.35), vegetation classification (1.28), hillshade (1.33), and the ruggedness index (1.25). The drop in average training gain with removal of distance from roads (1.13) eliminated the overlap in confidence intervals; therefore, the final model is comprised of north and east UTM, distance from streams, and distance from roads (Table 1). The AUC for the final four-variable model was 0.911. It predicted the distribution of summer GSG habitat about 3.5 times better than a random distribution, and was used to generate the map in Figure 4C. The reclassification procedure identified prediction values greater than 0.45 (11.3% of the study area) as suitable habitat.
Summer season without UTM coordinates
The AUC for the full model without north and east UTM was 0.89 and the average training gain was 1.08. The Jacknife test of variable importance (Figure 2) shows that the individual training gains for elevation (0.49), distance from streams (0.27), and vegetation classification (0.25), were highest among the set of predictor variables. Distance from streams lowered the average training gain the most when removed from the full model followed by elevation and then distance from roads.
Confidence intervals for the average training gain (0.99) with removal of aspect did not overlap with those of the full model, did overlap with the subsequent removal of hillshade (1.00), and finally did not overlap with the removal of curvature (0.965) and the remaining predictors. The training gain for the five-variable model containing elevation, distance from streams, vegetation classification, ruggedness index and distance from roads was chosen as the final model (Table 1), because it was not significantly different from the seven or six-variable models, hence it performed nearly same with fewer predictors. The final model predicted the distribution of summer GSG habitat about 2.6 times better than a random distribution and was used to generate the map in Figure 4D. The reclassification procedure identified prediction values greater than 0.36 (19.9% of the study area) as suitable habitat.
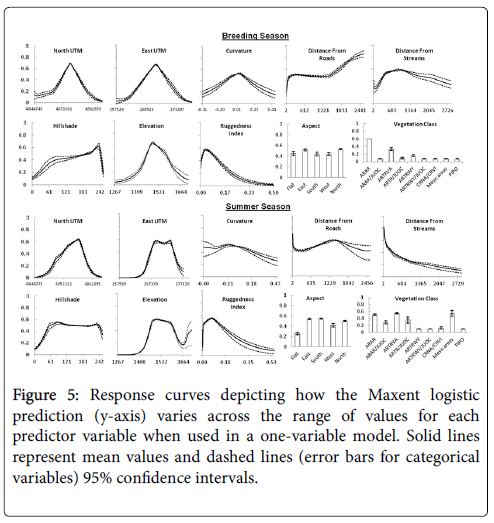
Response Curves
Response curves were generated to show how each predictor variable affects the Maxent prediction for models created using only the corresponding variable. These curves were also used to compare differences between the curves produced with breeding and summer season datasets for each predictor variable that entered one of the four final models (Figure 5). Average prediction values produced by east and north UTM from the breeding season data were symmetrically peaked near the middle of the range of both variables as might be expected given one location served as the center of the lek and the study area and that GSG congregate around leks during this season [8,21]. However, the peak was shifted to the right in the curve produced with summer dataset for North UTM and the highest prediction values were more spread out across east UTM.
The pattern of prediction values across the vegetation categories suggests GSG used habitat types relative to availability (Figures 2 and 5) during the breeding season except for the PIPO type in which no birds were recorded. Nonetheless, the percentage of relocations (75%) was disproportionately higher in the ARAR habitat type which comprised 34% of study area. The summer season dataset prediction values were notably higher for the mesic areas, ARAR/JUOC, ARTRVA, and ARTRVA/JUOC categories, reflecting different habitat utilization than during the breeding season. Maxent prediction values during the breeding season increased rapidly from about 0.3 for areas near streams and water sources, peaked at about 900 meters, and then gradually decreased to 0.32 at the uppermost range of distance values. The response curve produced with the summer dataset was considerably different with the highest prediction values associated with areas closest to streams followed by a steep curvilinear decrease in value with increasing distance from streams.
The response curve produced by elevation during the breeding season shows the highest prediction values peaked at mid-elevations of the study area and were near zero at the lowest and highest elevations. The summer dataset curve was similar except the peak was slightly lower in value and flattened across a larger range of elevation. Prediction values dropped as elevation increased but then jumped sharply for areas of highest elevation without a corresponding increase in frequency of GSG locations. This increase in the upper range of elevation values was likely caused by two birds located above 1,683 m elevation. Since the amount of area with at least this elevation represented only 0.2 percent of the study area these two location points likely inflated the habitat prediction values. The uncertainty in this prediction region of the variable is also higher given the wider confidence intervals.
Average prediction values were low for distances closest to roads but increased rapidly up to 200 m after which they stabilized until 1500 m when they steadily increased again with increasing distance. The response curve for the summer season dataset started with higher prediction values for areas closest to roads, increased to a peak around 1500 m and then decreased with further distance from roads. This decrease was accompanied by expanding confidence intervals however, indicating increasing uncertainty as distance from road increased. The responsive curve for ruggedness index was very similar for both datasets with average prediction values highest for areas with low terrain variation followed by a rapid decline as terrain variation increased.
Predictive Mapping
Maps produced by models with coordinates (Figures 4A and 4C) show a pattern of high prediction values concentrated over a relatively smaller area whereas maps produced by models without coordinates (Figures 4B and 4D) show a pattern of high prediction values spread out across a much larger part of the study area. The distribution of GSG locations during the breeding season were aggregated relatively close to the center of the study area and the model with coordinates produced a pattern of high prediction values that visually agrees with this aggregation (Figure 4A). However, a small number of locations closer to the outer boundary of the study area fell within areas with low prediction values. Absence of the vegetation cover class in the summer season model with UTM coordinates (Figure 4C) produced a smoother, less complex pattern of prediction values across the study area relative to the other maps produced from models with coordinates. This map shows how the predictor variable representing distance from streams influenced the model where the stream network was made more apparent than the other maps.
Discussion
We found that GSG routinely moved up to 10 km from winter and lekking areas to nesting and summering areas. They shifted from sites characterized by low juniper and high A. arbuscula dominated areas to sites with higher A. tridentata spp. vaseyana and A. arbuscula cover. We found these shifts in habitat use to be predictable across both space and time during the 3 years of our study. These movements and habitat choices across such large extents indicate that GSG management will be most effective when it incorporates actions across entire landscapes.
This study used the Maxent species distribution modeling software to generate habitat suitability models and distribution maps for the 2006-2008 breeding and summer seasons for a population of GSG. Response data were represented by telemetry locations of radio collared birds collected during this period of time and predictor variables were represented with a set of GIS-based biophysical attributes with a spatial extent limited to a 10 km radius centered at the middle of the GSG lek. This analysis provides empirically-mapped information to help biologists and managers understand the combined effects of landscape attributes that attract birds to preferred habitat during different seasons and predict the general directional movements of birds from breeding to summer seasonal habitats.
This research provides evidence that GSG selectively utilize habitats within broad landscapes. Based upon the models with and without UTM coordinates, 9.1% of the study area was estimated as suitable breeding habitat. The amount of suitable summer habitat estimated from the model with UTM coordinates was 11.3% of the study area and 19.9% estimated from the model without UTM coordinates. The amount of suitable breeding habitat that overlapped with suitable summer habitat was 1160 ha and 1457 ha for models with and without UTM coordinates, respectively. This supports the hypothesis developed by Kolasa and Waltho [33] that habitats, particularly for species using large areas, will usually be subunits embedded within the area they are using. Being able to predict suitable use areas can help managers allocate limited resources most effectively. For example, managers can prioritize areas for monitoring habitat suitability in meaningful locations versus random locations, and target areas for conservation and ecological rehabilitation/restoration actions within broad landscapes.
Models with UTM coordinates outperformed models without these variables and had nearly the same performance for both seasons. The higher capability to differentiate presences from background locations suggests that the set of environmental factors controlling habitat selection were not fully represented in the other predictor variables. Alternatively, the success of the coordinates suggests it might be the most useful landscape-scale information to account for the natural behavior of GSG to maintain a general aggregation near conspecifics during both seasons. Nonetheless, the region of high breeding habitat suitability identified with the model with spatial coordinates was also strongly coincident with a particular range of elevation values as reflected in the high correlation with values for East UTM, high training gain, and largest drop in training gain when removed from the model without UTM coordinates, for both seasons.
The A. arbuscula (ARAR-low sagebrush) cover type with less than 5% juniper cover had the highest predictive values during the breeding season, contained 75% of breeding season telemetry locations, and comprised the largest amount (34%) of the study area indicating higher proportion of use to availability. Others have reported similar findings of GSG selecting for low sagebrush communities during the pre-laying [34] and early brood-rearing [2] timeframes. In our study area, this cover type had greater annual forb cover (338%), a higher density of annual food forbs (174%), and we observed forb expression occurred earlier in the growing season than the average for the entire study area [35]. Increased annual forb availability and the early expression of forbs may explain why GSG selected the ARAR cover type during the breeding season as the consumption of forbs during the pre-laying period and early brood-rearing results in greater hen reproductive success and chick survival [2,34,35].
During the summer season the ARAR, ARTRVA, and Mesic Areas contained 48%, 40.5%, and 3.1% of telemetry locations which comprised 33%, 25%, and 1% of the study area, respectively, indicating a higher proportion of use to availability. Drut et al. [2] reported similar results in that GSG increased use of big sagebrush cover types later in the brood-rearing period and concentrated use near lakebeds and meadows. The ARTVA cover type (with less than 5% Juniper cover) had a greater cover of deep-rooted perennial grasses (107%), perennial forbs (160%), and annual forbs (200%) and greater perennial (113%) and annual (161%) food forbs than the average for the entire study area [35]. Having greater cover values and food forbs than surrounding vegetation communities may explain why GSG predicted use of this cover time is high during the summer season. GSG increased predicted use for Mesic Areas is similar to results reported by Dzialak et al. [36], who found that GSG selected for mesic habitat in mid and late brood-rearing phases. The Mesic Areas cover type had greater perennial forb cover (269%), perennial food for density (257%), and forbs remained succulent longer into the growing season than the average for the entire study area [35].
Even though the importance of the vegetation class to the Maxent models dropped with the seasonal transition it remained in the model without UTM coordinates and revealed significant changes among the cover types. During both the breeding and summer seasons, GSG preferred cover types with less than 5 % juniper canopy cover compared to those same cover types with greater than 5 % juniper canopy cover (Table 2).Similar to our results, Casazza et al. [37] found that at larger scales (7.9 – 226.8 ha) sage-grouse avoided pinyon pine and Utah juniper vegetation communities. Baruch-Mordo et al. [38] also found similar results in that at a low level of encroachment (>4% cover) no leks remained active suggesting GSG avoidance of Juniper. In our study, GSG during the summer season showed a notable increase in juniper habitat use during the breeding season. We did observe GSG shading under a single or small group of trees during the summer season; however, we did not differentiate between differing cover values (i.e., only > <5 percent canopy cover) or Juniper spatial configurations, which likely have an influence on GSG habitat use patterns [38].
| ARAR | ARAR JUOC | ARTRVA | ARTR JUOC | ARTRW | ARTRW JUOC | CHNA CHVI | Mesic | PIPO | |
|---|---|---|---|---|---|---|---|---|---|
| Area (%) | 33.0 | 7.0 | 25.0 | 5.0 | 14.0 | 1.0 | 3.0 | 1.0 | 10.0 |
| BreedingLocations (%) | 75.0 | 0.6 | 19.0 | 1.2 | 4.2 | 0.0 | 0.0 | 0.0 | 0.0 |
| SummerLocations (%) | 47.9 | 4.3 | 40.5 | 3.7 | 0.0 | 0.0 | 0.6 | 3.1 | 0.0 |
Table 2: Total percent of the area represented by each of the primary vegetation community types.
The low predictive values during the breeding season for the mesic areas subcategory within the vegetation classification variable along with the relatively low prediction values for distances close to streams (relative to the summer season) agrees with results reported by Yost et al. [13] who found that models of GSG nest habitat suitability in southcentral Oregon produced negative exponent values for riparian habitats. The indication GSG may have avoided mesic areas during the breeding season changed considerably during the summer season. Prediction values were much higher for the closest distances to streams and for the mesic areas subcategory within the vegetation classification. Results show that a general movement of GSG occurred from the center of the study area during the breeding season in a northeast direction to summer habitat. The shift in habitat was also reflected in high prediction values extended across a large range of elevation values where shrub cover types typically produce greater cover values. An increase in elevation is often associated with higher moisture availability to plants, particularly during the growing season when precipitation is limited and temperatures are higher [39,40]. Increased moisture availability at higher elevations is associated with increased vegetation cover and lengthens the time that forbs remain succulent. Similarly, Klebenow [1] found that GSG moved up in elevation following a gradient of green food plant availability in Idaho. In addition to succulent forb availability, a greater abundance of forbs that GSG consume was associated with increased elevation within the study area.
The distance from streams was a strong predictor variable that remained in models with and without UTM coordinates indicating a preference for green habitats during the summer. These results are consistent with Klebenow’s [1] observations of GSG that gathered near permanent water sources in late August where green vegetation remained succulent. Connelly et al. [20] also reported that GSG moved near agricultural land or more mesic habitats where succulent vegetation was likely to occur during the summer season. In addition to forbs remaining succulent through much of the summer in both perennial systems and meadows they also contained a greater density of GSG food forbs (150%) compared to all other cover types in the study area [35]. Drut et al. [2] reported an increased use of lakebeds and meadows during late brood-rearing (summer season) in southeastern Oregon. In addition to harboring succulent forbs longer into the summer season, Drut et al. [39] and Ersch [41] found that insect abundance increased in mesic habitats, which may attract hens and result in increased chick survival during early and late broodrearing periods [10]. Prediction values were relatively high for areas with gentle topography and diminished with increasing variation in terrain. These results are consistent with those reported by Dzialak et al. [37] who found GSG consistently avoided rough terrain.
Using species occurrence locations and ecological covariate data to estimate the seasonal changes in the distribution of a species provides the opportunity of conducting studies in GSG conservation and distribution that are difficult or impossible using other methods. However, we are aware that inferences from the models produced from this study are only as good as the models themselves and which can only be abstractions of the species fundamental niche [14]. The ecological characteristics of species affects model accuracy potential, where species widespread in both geographic and environmental space are generally more difficult to model than species with compact spatial distributions [42], as was the case with the sage-grouse data. The results of this study must also be considered in the context of the list of serious pitfalls that could affect the accuracy of predictive modeling and mapping with occurrence data described by Phillips et al. [24] and Yackulic et al. [43]. For example, all occurrence localities have some level of precision or error, can be biased by access conduits, sampling barriers, and variation in sampling effort over space and time. This analysis was based on presence-only data comprised of repeat telemetry locations of a small number of individuals from a larger population and results would most likely vary from what is reported here had different individuals been captured and tracked.
The choice of variables to use for building models directly affects the degree to which a model can be generalized to other areas and time periods. The set of modeling variables might be insufficient to describe all the parameters of a species fundamental niche relevant to its distribution at the grain of the modeling task. Large errors within the predictor variables will directly affect model accuracy. We recognize that the models reported as final through the variable selection process used might have included more predictor variables had we used a higher number of replicates effectively reducing confidence intervals for training gain values leading to less overlap among competing reduced models. The choice of modeling features and different parameter settings available for calibrating models with the Maxent software influences the results that can be achieved with a particular dataset. Results of this study were influenced in large part by the spatial extent of the vegetation class GIS layer which excluded telemetry locations outside the 10 km radius from contributing to the models.
We used the threshold value of P/E = 1 to objectively identify a prediction value for reclassifying the continuous logistic output to a binary map [22,32] of suitable/non-suitable habitat. However, we found that the choice of a larger width than what was used in this study for interpolating the values comprising the ratio results in smaller confidence intervals and a lower reclassification value, whereas the choice of smaller widths, just the opposite. For instance, output width values of 0.05 to 0.25 resulted in reclassification values of 0.6 to 0.23, respectively, a difference of 16% of the study area. Therefore, use of the P/E ratio with Maxent output requires a proviso that the amount of area allocated to suitability classes depends on the width of output values chosen to calculate the ratio. Use of the average from the results of a range of output widths might be a way to more objectively identify the prediction values to reclassify Maxent output into discrete habitat suitability classes [44]. These results are similar to those reported by Aldridge and Boyce [16] who found that nesting females had a strong avoidance of anthropogenic habitats such as roads.
Implications
Management agencies are required to work with limited resources relative to the extensive landscapes they typically manage. GSG selectively utilize portions within broad landscapes; therefore, managers can target areas for protection and conservation actions (ecological rehabilitation/restoration) stretching limited resources further. This requires that managers assess the landscape attributes that drive GSG habitat use. Managers can use this information to more effectively implement habitat restoration actions in meaningful locations.
Replication and more research is needed to refine and improve the predictive ability in other GSG habitat areas. Although general trends from this research may help managers predict GSG movement patterns and subsequently target areas for conservation emphasis, managers would benefit from greater improvements in modeling that identify specific areas that GSG are likely to utilize. Supplemental information addressing the influence of patch and resource spatial distribution across landscapes on GSG population sustainability would further augment conservation efforts.
Acknowledgement
We would like to thank Jennifer Bruce for her assistance with trapping birds and collecting data. We also thank Christian Hagan, George Buckner, Jan Hanf, and Matt Obradovich for their contribution in study design, logistics, and funding. We appreciate the support of Oregon State University (Department of Rangeland Ecology and Management) in Corvallis Oregon, Brigham Young University (Department of Plant and Wildlife Sciences) in Provo, Utah, the United States Bureau of Land Management of Oregon, and the Oregon Department of Fisheries and Wildlife.
References
- Klebenow DA (1969) Sage grouse nesting and brood habitat in Idaho. J WildlManag 33: 649-662.
- Drut MS, Crawford JA, Gregg MA (1994) Brood habitat use by sage grouse in Oregon.Great Basin Naturalist 54: 170-76.
- Gregg MA, Crawford JA, Drut MS, DeLong AK (1994) Vegetation cover and predation of sage grouse nests in Oregon. J WildlManag 58: 162-166.
- Delong AK, Crawford JA, Delong DC (1995) Relationships between vegetational structure and predation of artificial sage grouse nests. J WildlManag 59: 88-92.
- Sveum CM, Edge WD, Crawford JA (1998) Nesting habitat selection by sage grouse in south-central Washington.Journal of Range Management 51: 265-269.
- Doherty KE, Naugle DD, Walker BL, Graham JM (2010) Greater sage-grouse winter habitat selection and energy development. J WildlManag 71: 187-195.
- Aldridge CL, Saher DJ, Childers TM, Stahlnecker KE, Bowen ZH (2011) Crucial nesting habitat for Gunnison sage-grouse:a spatially explicit hierarchical approach.J WildlManag 76: 391-406.
- Connelly JW, Schroeder MA, Sands AR, Braun CE (2000)Guidelines to manage sage grouse populations and their habitats. Wildlife Society Bulletin 28: 967-985.
- Leonard KM, Reese KP, Connelly JW (2000) Distribution movements and habitats of sage grouse Centrocercusurophasianus on the upper Snake River plain of Idaho: changes from the 1950s to the 1990s.Wildlife Biology 6: 265-270.
- Gregg MA (2006) Greater sage-grouse reproductive ecology: linkages among habitat resources, maternal nutrition, and chick survival.Dissertation, Oregon State University, Corvallis, USA.
- Miller RF, Eddleman LL (2001) Spatial and temporal changes of sage grouse habitat in the sagebrush biome.Oregon Agricultural Experiment Station. Burns, Oregon, USA.
- Fuhlendorf SD, Woodward AJW, Leslie DM, Shackford JS (2002) Multi-scale effects of habitat loss and fragmentation on lesser prairie-chicken populations of the U.S. southern Great Plains.Landscape Ecology 17: 617-628.
- Yost AC, Petersen SL, Gregg MA, Miller RF (2008) Predictive modeling and mapping sage grouse (Centrocercusurophasianus) nesting habitat using Maximum Entropy and a long-term dataset from southern Oregon.Ecological Informatics 3: 375-386.
- Warren DL, Seifert SN (2011) Ecological niche modeling in Maxent: the importance of model complexity and the performance of model selection criteria. Ecological Applications 21: 335-342.
- Braun EC, Britt T, Wallestad RO (1977) Guidelines for maintenance of sage grouse habitats.Wildlife Society Bulletin 5: 99-105.
- Aldridge CL, Boyce MS (2007) Linking occurrence and fitness to persistence: A habitat-based approach for endangered greater sage-grouse. Ecological Applications 17: 508-526.
- Bruce JR, Robinson WD, Petersen SL, Miller RF (2011) Greater sage-grouse movements and habitat use during winter in central Oregon. Western North American Naturalist 71: 418-424.
- Anderson EW, Borman MM, Krueger WC (1998) The ecological provinces of Oregon: a treatise on the basic ecological geography of the state. Oregon Agricultural Experiment Station. Burns, Oregon, USA.
- Giesen KM, Shoenberg TJ, Braun CE (1982) Methods for trapping sage-grouse in Colorado.Wildlife Society Bulletin 10: 224-231.
- Connelly JW, Browers HW, Gates RJ (1988) Seasonal movements of sage grouse in southeastern Idaho. J WildlManag 52: 116-122.
- Crawford JA, Olson RA, West NE, Mosley JC, Schroeder MA, et al. (2004) Ecology and management of sage-grouse and sage-grouse habitat.J Range Manage 57: 2-19.
- Davis RJ, Dugger KM, Mohoric S, Evers L, Aney WC (2011) Northwest Forest Plan��?the first fifteen years (1994-2008): status and trends of northern spotted owl populations and habitats.Gen. Tech. Rep. PNW-GTR-850.Portland, OR: U.S. Department of Agriculture, Forest Service, Pacific Northwest Research Station. USA. pp: 147.
- Merow C, Smith MJ, Silander JA (2013) A practical guide to MaxEnt for modeling species��? distributions: what it does, and why inputs and settings matter. Ecography 36: 1058-1069.
- Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecological Modeling 190: 231-259.
- Elith J, Phillips SJ, Hastie T, Dudik M, Chee YE, et al. (2011) A statistical explanation of MaxEnt for ecologists. Diversity and Distributions 17: 43-57.
- Sappington JM, Longshore KM, Thompson DB (2007) Quantifying landscape ruggedness for animal habitat analysis: a case study using bighorn sheep in the Mojave Desert.J WildlManag 71: 1419-1426.
- Landis JR, Koch GG (1977) The measurement of observer agreement for categorical data. Biometrics 33: 159-174.
- Lobo JM, Jimenez-Valverde A, Real R (2007) AUC: a misleading measure of the performance of predictive distribution models.Global Ecology and Biogeography. pp: 7.
- Fielding AH, Bell JF (1997) A review of methods for the assessment of prediction errors in conservation presence/absence models. Environmental Conservation 24: 38-49.
- Swets JA (1988) Measuring the accuracy of diagnostic systems. Science 240: 1285-1293.
- Boyce MS, Vernier PR, Nielsen SE, Schmiegelow FKA (2002) Evaluating resource selection functions. Ecological Modelling 157: 281-300.
- Hirzel AH, Le Lay G, Helfer V, Randin C, Guisan A (2006) Evaluating the ability of habitat suitability models to predict species presences. Ecological Modelling 199: 142-152.
- Kolasa J, Waltho N (1998) A hierarchical view of habitat and its relationship to species abundance. In: Peterson DL. Parker VT (eds) Ecological scale: theory and applications. Columbia University Press, New York, NY.pp: 55-78.
- Barnett JK, Crawford JA (1994) Pre-laying nutrition of sage grouse hens in Oregon. JRange Manage 47: 114-118.
- Freese MT (2009) Linking greater sage-grouse habitat use and suitability across spatiotemporal scales in central Oregon.Thesis, Oregon State University, Corvallis, USA.
- Dzialak MR, Olson CV, Harju SM, Webb SL, Mudd JP, et al. (2011) Identifying and Prioritizing Greater Sage-Grouse Nesting and Brood-Rearing Habitat for Conservation in Human-Modified Landscapes. PLoS ONE 6: e26273.
- Casazza ML, Coates PS, Overton CT (2011) Linking habitat selection and brood success in greater sage-grouse. Studies in Avian Biology 39: 151-167.
- Barunch-Mordo S, Evans JS, Severson JP, Naugle DE, Maestas JD, et al. (2013) Saving sage-grouse from the trees: a proactive solution to reducing a key threat to a candidate species. Biological Conservation 167: 233-241.
- Drut MS, Pyle WH, Crawford JA (1994) Technical note: diets and food selection of sage grouse chicks in Oregon. J Range Manage 47: 90-93.
- Swanson FJ, Kratz TK, Caine N, Woodmansee RG (1988) Landform effects on ecosystem patterns and processes. BioScience 38: 992-98.
- Ersch EA (2009) Effects of plant community characteristics on insect abundance: Implications for sage-grouse brood-rearing habitat. Thesis, Oregon State University, Corvallis, USA.
- Hernandez PA, Graham CH, Master LL, Albert DL (2006) The effect of sample size and species characteristics on performance of different species distribution modeling methods. Ecography 29: 773-785.
- Yackulic CB, Chandler R, Zipkin EF, Royle A, Nichols JD, et al. (2012) Presence-only modeling using MAXENT: when can we trust the inferences?Methods in Ecology and Evolution 4: 236:243.
- Renner IW, Warton DI (2013) Equivalence of MAXENT and Poisson point process models for species distribution modeling in ecology.Biometrics 69: 274-281.
Relevant Topics
- Aquatic Ecosystems
- Biodiversity
- Conservation Biology
- Coral Reef Ecology
- Distribution Aggregation
- Ecology and Migration of Animal
- Ecosystem Service
- Ecosystem-Level Measuring
- Endangered Species
- Environmental Tourism
- Forest Biome
- Lake Circulation
- Leaf Morphology
- Marine Conservation
- Marine Ecosystems
- Phytoplankton Abundance
- Population Dyanamics
- Semiarid Ecosystem Soil Properties
- Spatial Distribution
- Species Composition
- Species Rarity
- Sustainability Dynamics
- Sustainable Forest Management
- Tropical Aquaculture
- Tropical Ecosystems
Recommended Journals
Article Tools
Article Usage
- Total views: 11661
- [From(publication date):
September-2016 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 10750
- PDF downloads : 911