Research Article Open Access
Selected Reaction Monitoring: A Valid Tool for Targeted Quantitation of Protein Biomarker Discovery
Subodh Kumar* and Priyanka Mittal
Central Research Laboratory, Multi-disciplinary Research Unit, University College of Medical Sciences (University of Delhi) and GTB Hospital, Delhi, India
- *Corresponding Author:
- Subodh Kumar
Research Scientist, Multi-Disciplinary Research Unit
UCMS and GTB Hospital
University of Delhi, Delhi-110 095, India
Tel: +911122592977
E-mail: subodh_bt2003@yahoo.co.in
Received Date: April 14, 2017 Accepted Date: May 08, 2017 Published Date: May 12, 2017
Citation: Kumar S, Mittal P (2017) Selected Reaction Monitoring: A Valid Tool for Targeted Quantitation of Protein Biomarker Discovery. J Anal Bioanal Tech 8: 362. doi: 10.4172/2155-9872.1000362
Copyright: © 2017 Kumar S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
Biomarker discovery is relying on the sensitivity and specificity of the detected biomolecules in clinical samples. Several potential biomarkers against the particular disease cannot be detected because of the unavailability of either specific procedure or sensitive instrument or both, which hamper the diagnosis. In spite of that some of the potential biomarkers are available in trace amount in biological fluids like serum, urine, buccal swab, sputum etc. and need a sensitive method to detect precisely those biomolecules. Now days Selected Reaction Monitoring (SRM) is Mass Spectrometry based approached who can overcome the problems associated with biomarker discovery. India have adequate and variety of clinical samples but some time due to lack of infrastructure and knowledge we are unable to utilise those samples for human welfare. In this review, we are discussing about the experimental setup and procedure of SRM experiment.
Keywords
LC-MS/MS; Triple quad; Selected reaction monitoring; Biomarker; Diagnosis
Abbreviations
LC-MS: Liquid Chromatography-Mass Spectrometry; SRM: Selected Reaction Monitoring; ELISA: Enzyme Linked Immuno-Sorbent Assay; ICT: Immunochromatography Test; SDS-PAGE: Sodium do-decyl sulphate-Poly acrylamide gel electrophoresis; DTT: Dithiothreitol; IAA: Indole Acetic Acid; MS: Mass Spectrometer; ESI: Electro Spray Ionization.
Introduction
Mass spectrometry (MS) is an analytical technique which is used to measure sample’s molecular mass by measuring mass to charge ratio. In this method, a soft ionization technique like ESI (Electro Spray Ionization) is used to generate charge ions. Now days, this is the most popular method for protein identification and quantitation. In shotgun proteomics proteins are digested into smaller peptides followed by analysis of complex mixture of peptides on high performance liquid chromatography (HPLC) coupled with mass spectrometry. MS based proteomics approach can be designed to use either nontargeted (shotgun) or targeted (SRM) proteomics. Bottom up shotgun MS approach is commonly used in discovery proteomics. Shotgun proteomics is a powerful tool for high-throughput peptide/protein identification and relative quantification but unsuitable for absolute quantification. In contrast to this, MS based targeted proteomics is mostly used for absolute quantification of small set of protein or peptide.
Targeted Proteomics
In contrast to shotgun proteomics [1], targeted proteomics is now days play a very important role in the accurate measurement of protein of interest in complex biological sample which can be helpful in dissecting the protein network and pathway. Selected Reaction Monitoring (SRM) is one such approach.
SRM-MS (Selected Reaction Monitoring-Mass Spectrometry)
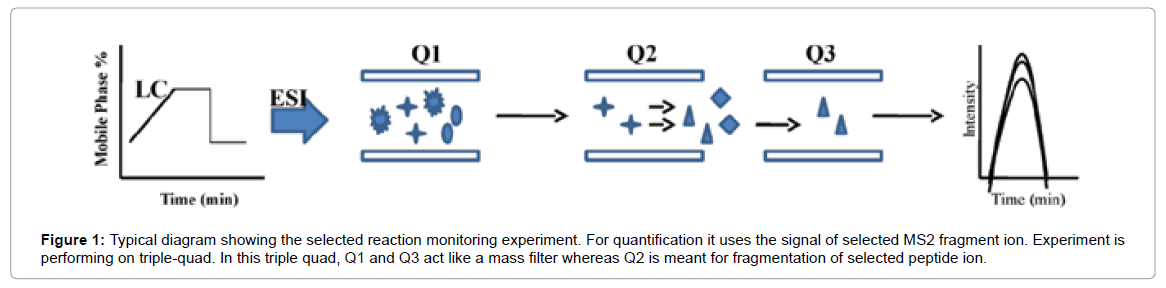
Selected Reaction Monitoring (SRM) is a non-scanning technique, generally performed on triple quadrupole (QQQ) instruments in which fragmentation is used as a means to increase selectivity. In SRM, the first and third quadrupoles act as filters to specifically select predefined m/z values corresponding to the peptide ion and a specific fragment ion of the peptide, whereas the second quadrupole serves as collision cell (Figure 1).
Figure 1: Typical diagram showing the selected reaction monitoring experiment. For quantification it uses the signal of selected MS2 fragment ion. Experiment is performing on triple-quad. In this triple quad, Q1 and Q3 act like a mass filter whereas Q2 is meant for fragmentation of selected peptide ion.
SRM is a revolutionary tool for clinical application, for biomarker validation of blood plasma [2]. The method proposed in this review is a targeted proteomics approach which combines multiple approaches investigating a certain set of proteins in more detail. One such targeted proteomics approach is the combination of liquid chromatography and selected reaction monitoring or multiple reactions monitoring (SRM/MRM). The robustness of this methodology not only allows for consistent measurement of different sample conditions, but also obtains reliable results across various laboratories with coefficient of variation less than 20% [3]. SRM-MS requires the prior knowledge of fragmentation pattern of the targeted peptide because the analytes in a sample is determined by measuring m/z values of predefined precursor and fragment ions.
The two level of mass selection with narrow mass windows results in a high selectivity as co-eluting background ions are filtered out very effectively. These dual filters provide a higher sensitivity for monitoring specific analytes as supposed to conventional shotgun experiment. The set of precursors and their corresponding fragment ion is usually referred as “TRANSITIONS”. Unlike in other MS-based proteomics techniques, no full mass spectra are recorded in QqQbased SRM analysis resulting in an increased sensitivity by one or two orders of magnitude compared with conventional full scan techniques. Typically, positive detection of specific precursor/peptide of interest is usually based on the positive detection of atleast three to five coeluting transition ions of particular precursor/peptide of interest, hence, SRMMS does not depend on single spectrum for positive identification, but on coeluting transitions [4]. In an unscheduled MRM, the number of transitions is defined by the dwell time and cycle time. The dwell time is the time to take each transition and cycle time is the times to complete all the transitions listed in the methods. The higher the dwell-time, the higher the signal-to-noise ratio and thus lower the limit of detection [5,6]. Therefore, it’s very important to set the dwell time 2-4 minutes time window for specific and reproducible detection for peptide/ protein of interest. In spite of the dwell and cycle time, some instrument specific set up time is also considered that switch the m/z of the filter. For example if each transition is measured for 30 ms dwell time and instrument set up time is 3 ms, in 2000 ms approximately 60 transitions can be measured. In an unscheduled SRM-MS, only 12 peptides can be measured with 5 transitions per peptide. Since the numbers of peptide detected is low therefore scheduled SRM-MS could be a choice but the limitation with this method is that we should know the retention time of the peptide. If we wish to effectively monitor hundreds of assays in a single injection during LC-SRM-MS run, the window should be set between 2-4 minutes.
In the proposed review we discussed in brief about the instrumentation, parameters etc. of targeted proteomics approach for biomarker discovery.
General procedure to establish a proteomic SRM experiment
In contrast to the conventional shotgun proteomic studies, in LC-SRM-MS, three main selection processes should be kept in mind before going to start validation of the experiment and these are; first the information about the protein that is to be targeted; second, the selection of those peptide that present good MS response and uniquely identify the targeted protein, such peptide are known as proteotypic peptide (PTPs) [7] and third, identified those PTP (Proteotypic peptide) who have optimal signal intensity. The overall workflow of the experiment is mentioned in Figure 2. The major steps that are involved in the experiment are explained in the following sections.
Figure 2: Typical workflow of a Selected Reaction Monitoring (SRM) experiment. It is typically performed on triple quadrupole instruments. Q1 and Q3 acts like a mass filter whereas Q2serves as a collision cell. Q1 select a peptide ion, Q2 fragment the selected peptide ion and Q3 selects a specific fragment ion for detector. The double mass selection reduces possible interferences between ions.
Selection of target protein
In LC-SRM-MS, the researcher should have prior knowledge of protein and their peptide behaviours which includes the retention time and fragmentation pattern. The major issue is with the selection of those target proteins which are present in low abundance in a complex sample. This information is either present in the repositories data base available online or have to be determined before SRM-MS experiment through bottom up shotgun LC-MS/MS experiment. Several, sites are available freely to select the target proteins (Table 1a) [8-14]. The success of shotgun proteomics relies on the sample preparation. In this section, we are discussing about the proteomics sample preparation.
| S No | Software | Link | References |
|---|---|---|---|
| 1 | Gene expression GEO | [8] | |
| 2 | Protein expression Protein Atlas | [9] | |
| 3 | Gene ontology groups GO | [10] | |
| 4 | Functional groups KEGG | [11] | |
| 5 | Protein–protein interactions IntAct | [12] | |
| 6 | Protein–protein interactions MINT | [13] | |
| 7 | Network expansion PhosphoPep | [14] |
Table 1a: Online information resources relevant to selection of a set of proteins of interest [4].
Sample preparation for shotgun proteomics: Add four volume of chilled acetone to one volume of plasma/serum sample followed by vortex for complete breakdown. Store the sample at -20°C for overnight. After incubation, spin and the resulting pellet should wash away with chilled acetone. Add one volume of buffer containing 8 M urea in 100 mM ammonium bicarbonate (pH 8.0) to initial volume of the sample. Subsequently add protease and phosphatase inhibitor in the sample. Vortex thoroughly and centrifuge the dissolve protein. Collect the supernatant in a fresh tube and do the protein estimation either through Lowry or BCA method. Dilute sample to 1 M urea concentration with 100 mM ABB buffer followed by reduction (with 20 mM DTT, at 60°C for 20-30 min) and alkylation (with 60 mM IAA, in dark for 30 minutes). The resulting sample will digest two times with trypsin at 3-4 hrs interval followed by overnight incubation at 37°C incubator. The reaction will be stop by adding 10% acetic acid to the digested sample. The digested sample desalt with C18 cartridge. The desalted samples now do the fractionation with either 1D or 2D separation on HPLC and collect the fractionated samples. Speedvac the samples to dry in vacuum and reconstitute with the appropriate solvent. The reconstituted sample will be vortex vigorously followed by centrifugation to collect the upper layer of the sample. Now the sample is ready to inject in mass spectrometer for proteomic study.
In gel digestion: The SDS-PAGE gel containing desired protein band will be place in petridish and fill it with MilliQ water. Remove the gel from pertidish and place it on glass slide. Cut the gel into their respective bands and dice the gel slice into uniform pieces and place them in eppendorf tube. The tube containing dice piece of gel is soaked in 0.5 ml of 25 mM ABB (Ammonium Bicarbonate) buffer and vortex for 10-15 min. To destain the gel, add 1 ml of 25 mM ABB buffer containing 50% ACN. Vortex for 20-30 minutes at RT (Room Temperature) followed by several washing. Dehydrate the gel with 100% ACN (Acetonitrile) solution by vortexing. For reduction and alkylation incubate the tube with 10 mM DTT (for 1 h at 56°C in water bath) and 55 mM IAA (for 45 minutes at RT in dark), respectively. Digest the gel with freshly prepare trypsin solution. Put on ice for 15 minutes until the gel particles absorb the buffer completely. Transfer gel to 37°C and incubate overnight. Finally the peptides will extract by adding 2 volume of 50% ACN containing 0.1% TFA. Vortex and collect the supernatant in a fresh tube and speed vac to dry the supernatant in vaccum. Desalt the extracted peptide on C18 cartridge. After desalting, dissolve the peptide in appropriate solvent for LC-MS/MS analysis.
Peptide selection
Upon trypsin digestion, each protein generates ten to hundreds of peptides [15]. The sensitivity of the MRM assay relies on the selection of peptides with favourable mass spectrometry properties and these are;
MS properties: A number of peptide generates after digestion with trypsin but only a small subset is routinely observed because they satisfy the required MS properties [16]. For assay development, the time can be reduced significantly if previous information generated from multiple shotgun experiment, deposited in online repositories is used to select those peptide that are most likely observed in the experiment and provide the strongest specific signals (Table 1a).
Uniqueness: For targeted MS analysis, it is essential to ensure that the peptide selected uniquely identify the targeted protein or one isoform thereof.
Post Translational Modification (PTM): Modified peptide can’t be detected by SRM unless specifically targeted. For reliable quantification, at least two should be monitored for each targeted protein.
Chemically induced modification: The potential source of error in quantitative MS experiments, during sample processing is the introduction of artifactual chemical modification in the sequence. Therefore, avoid those peptides with a high propensity for artifactual modifications. Generally, avoid peptides containing Methionine or Tryptophan and Glutamine or Asparagine in the sequence, as they are prone to oxidation and deamidation, respectively.
Cleavage site: Data analysis in shotgun proteomics experiment revealed a number of missed cleavage due to incomplete digestion of trypsin. These peptide sequences should be avoided for absolute quantification experiment. In general, peptide with two neighbouring basic amino acids at either cleavage site of the protein sequence should be avoid as those sites are predestined for a high rate of missed cleavage.
Selection of SRM Transitions
In general, for SRM experiment per peptide atleast 2-3 transition will be selected whose intensity is comparatively more so that it can be quantitate effectively. There is commercially available software which generates peptide transition in-silico. The commercially available software that helps in the selection of peptide transitions is listed in Table 1b [17,18].
| S No | Software | Link | Vendor/Lab | Reference |
|---|---|---|---|---|
| 1 | MRmaid | Bessant/Cranfield University | [17] | |
| 2 | MRMPilot | AB Sciex | Commercial | |
| 3 | Pinpoint | Thermo Scientific | Commercial | |
| 4 | Skyline | MacCoss Lab, U Washington | [18] |
Table 1b: List of software’s uses for selected Reaction Monitoring (SRM) Experiment’s development.
Validation of transitions
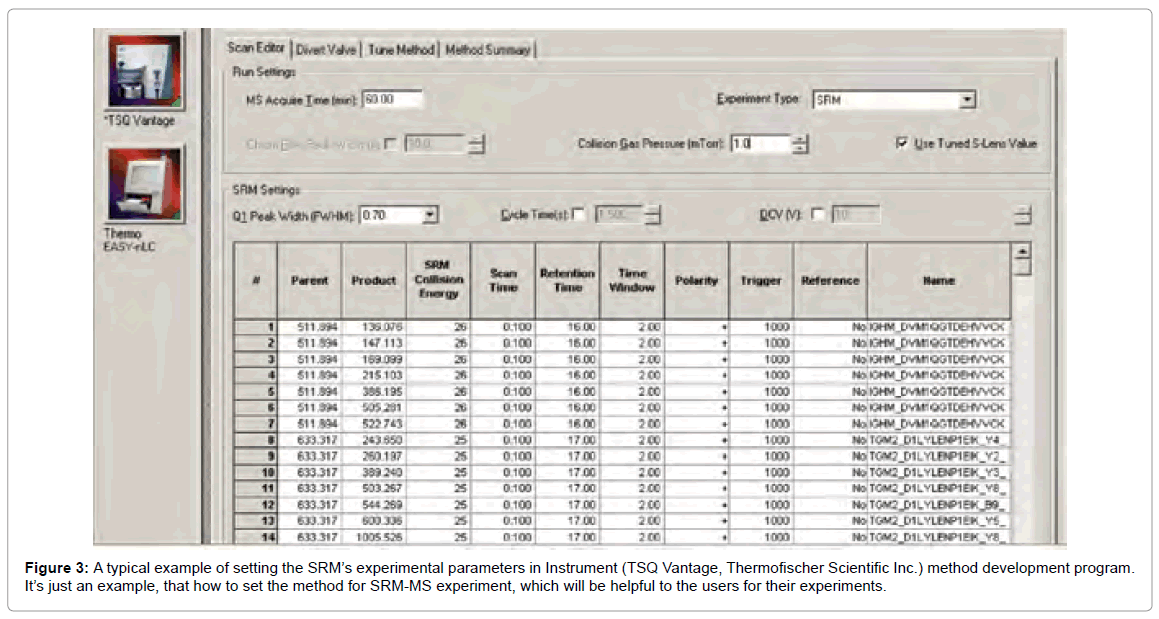
Peptides with precursor/fragment ion pairs of similar masses shows unspecific signal. This is because of the closely related sequences so that parts of the transitions are identical. The first step of validation is the parallel acquisition of several transitions for a targeted peptide (Figure 3). At the time of peptide elution, such transition yield a perfect set of coeluting intensity peaks if they are derived from same peptide.
Quantification of SRM transitions
Two emerging core strategies for targeted biomarker quantitation are
1) LC-SID-MRM-MS and
2) The combination of the so called SISCAPA technology.
The gold standard for MS-based quantitative method is to use stable-isotope-labelled peptide standard spiked into the sample of interest which then allow quantitation from a simple ratio of the labelled peak height or peak area to the unlabelled. However, some analogue IS can also work well when stable isotope labelled IS is not available. This stable isotope dilution (SID) method is the basis for SRM or MRM method. When some of the peptides are spiked with isotopic reference peptides, the corresponding endogenous peptides with the same sequences can be accurately quantified.
Discussion
An ideal biomarker has high sensitivity and specificity against a particular state of the disease and it should be validated on atleast 200 control group samples. On the other hand antigen based diagnosis is more advantageous over the antibody based detection. The major drawback of antibody based detection is their cross reactivity with other similar diseases, showing false positivity with endemic healthy controls and unable to differentiate between past and active infection etc. especially in infectious diseases. In infectious diseases, during active infection, several host or parasite specific proteins either up regulate or down regulate and these proteins can be a biomarker for the particular disease, who can overcome the above mentioned problem but the limitation is their detection and quantitation in clinical samples as they are present in trace amount. On the other hand, in non-communicable diseases like cancer, atherosclerosis, diabetes etc. several diagnostic biomarkers had been identified in the past by using mass spectrometry based proteomics approach [19-21] but their quantitative estimation in real samples had not been followed. Now days, targeted proteomics approach of triple quadrupole based Selected Reaction Monitoring (SRM) is experiment of choice which is able to quantitate effectively and sensitively in clinical sample. In this review, we have discussed about the importance and experimental procedures which will be helpful in designing the biomarker discovery experiment and will be helpful in achieving the goal of elimination program of communicable and non-communicable diseases.
Conclusion
SRM is a revolutionary MS based approach for a confident quantitation of small set of proteins/peptides in complex biological samples. This method is highly sensitive and specific as double mass selection reduces the possible Ion interference. Previously, due to lack of such a sensitive technique we were unable to analyse and validate the known biomarker against a specific disease across different set of samples. This analytical based method will surely be helpful in near future in improving the human health by identifying those biomarkers which is either present is trace amount or differentially express. After the successful identification, in long term goal, from the final outcome, we can develop simple and easy detection format, either in the form of Immunochromatographic strip (ICT) or ELISA (Enzyme Linked Immunosorbent Assay) format for commercial use. Therefore, we can utilize this technique for biomarker discovery against several deadly diseases to support the elimination program.
Conflict of Interest
The authors declare that there is no conflict of interest.
Acknowledgements
Author would like thanks to Department of Health Research-Indian Council of Medical Research, New Delhi, India and Central Research Laboratory, UCMS and GTB Hospital for their support to complete this review. Author would also like thanks to Dr. SK Sze, Newman, Nanyang Technological University, Singapore, for their guidance in understanding the selected reaction monitoring (SRM) experiment.
References
- Hayes McDonald W, John Yates R (2002) Shotgun proteomics and biomarker discovery. Disease Markers 18: 99-105.
- Anderson L, Hunter CL (2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics 5: 573-588.
- Prakash A, Tomazela DM, Frewen B (2009) Expediting the development of targeted SRM assays: using data from shotgun proteomics to automate method development. J Proteome Res 8: 2733-2739.
- Barnidge DR, Dratz EA, Martin T (2003) Absolute quantification of the G protein-coupled receptor rhodopsin by LC/MS/MS using proteolysis product peptides and synthetic peptide standards. Anal Chem 75: 445-451.
- Lange V, Picotti P, Domon B (2008) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol 4: 222.
- Picotti P, Aebersold R (2012) Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods 9: 555-566.
- Mallick P, Schirle M, Chen SS (2007) Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol 25: 125-131.
- Barrett T, Troup DB, Wilhite SE (2007) NCBI GEO: mining tens of millions of expression profiles database and tools update. Nucl Acids Res 35: D760-D765.
- Uhlen M, Ponten F (2005) Antibody-based proteomics for human tissue profiling. Mol Cell Proteomics 4: 384-393.
- Ashburner M, Ball CA, Blake JA (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25-29.
- Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucl Acids Res 28: 27-30.
- Kerrien S, Alam-Faruque Y, Aranda B (2007) IntAct-open source resource for molecular interaction data. Nucl Acids Res 35: D561-D565.
- Chatraryamontri A, Ceol A, Palazzi LM (2007) MINT: The Molecular INTeraction database. Nucl Acids Res 35: D572-D574.
- Bodenmiller B, Malmstrom J, Gerrits B (2007) PhosphoPep–a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol Syst Biol 3: 139.
- Picotti P, Aebersold R, Domon B (2007) The implications of proteolytic background for shotgun proteomics. Mol Cell Proteomics 6: 1589-1598.
- Kuster B, Schirle M, Mallick P (2005) Scoring proteomes with proteotypic peptide probes. Nat Rev Mol Cell Biol 6: 577-583.
- Mead JA, Bianco L, Ottone V (2009) MR Maid, the web-based tool for designing multiple reaction monitoring (MRM) transitions. Mol Cell Proteomics 8: 696-705.
- MacLean B, Tomazela DM, Shulman N (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Gene expression 26: 966-968.
- Prieto DA, Johann DJ, Bih-Rong W (2014) Mass spectrometry in cancer biomarker research: a case for immuno depletion of abundant blood-derived proteins from clinical tissue specimens. Biomark Med 8: 269-286.
- Tessitore A, Gaggiano A, Cicciarelli G (2013) Serum Biomarkers Identification by Mass Spectrometry in High-Mortality Tumors. International Journal of Proteomics.
- Dang VT, Werstuck GH (2016) Metabolomics - Based Biomarkers of the Pathogenesis of Atherosclerosis. Biomark J 2: 10.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 4749
- [From(publication date):
June-2017 - Nov 21, 2024] - Breakdown by view type
- HTML page views : 3996
- PDF downloads : 753