Research Article Open Access
Search for Low Molecular Weight Compounds that Inhibit Human Immunodeficiency Virus Type 1 Replication
| Chris Verathamjamras1#, Yu-Shi Tian2#, Norihito Kawashita2,3, Kousuke Okamoto2, Teruo Yasunaga3, Kazuyoshi Ikuta3, Kazushi Motomura1,3, Naokazu Takeda1,3, Tatsuya Takagi2,3* and Masanori Kameoka1,3,4* | ||
| 1Thailand-Japan Research Collaboration Center on Emerging and Re-emerging Infections (RCC-ERI), Building 10, Department of Medical Sciences, Ministry of Public Health, Muang, Nonthaburi 11000, Thailand | ||
| 2Graduate School of Pharmaceutical Sciences, Osaka University, Suita, Osaka 565-0871, Japan | ||
| 3Research Institute for Microbial Diseases, Osaka University, Suita, Osaka 565-0871, Japan | ||
| 4Division of Infectious Diseases, Department of International Health, Kobe University Graduate School of Health Sciences, Kobe, Hyogo 654-0142, Japan | ||
| #These authors equally contributed to this work. | ||
| Corresponding Authors : | Masanori Kameoka Division of Infectious Diseases, Department of International Health Kobe University Graduate School of Health Sciences, Kobe, Hyogo 654-0142, Japan Tel: +81 78 7964594 Fax: +81 78 7964594 E-mail: mkameoka@port.kobe-u.ac.jp |
|
| Tatsuya Takagi Graduate School of Pharmaceutical Sciences Osaka University, 1-6 Yamadaoka, Suita, Osaka 565-0871, Japan Tel: +81 6 68798244 Fax: +81 6 68798242 E-mail: satan@gen-info.osaka-u.ac.jp |
||
| Received April 16, 2015; Accepted May 20, 2015; Published May 27, 2015 | ||
| Citation: Verathamjamras C, Tian Y, Kawashita N, Okamoto K, Yasunaga T, et al. (2015) Search for Low Molecular Weight Compounds that Inhibit Human Immunodeficiency Virus Type 1 Replication. J Infect Dis Ther 3:218. doi: 10.4172/2332-0877.1000218 | ||
| Copyright: © 2015 Verathamjamras C et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | ||
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
||
Visit for more related articles at Journal of Infectious Diseases & Therapy
Abstract
Highly active antiretroviral (ARV) therapy has successfully reduced viral transmission, morbidity, and mortality associated with human immunodeficiency virus type 1 (HIV-1) disease; however, the emergence of drug-resistant viruses is a major obstacle associated with ARV therapy. Therefore, the development of a new class of ARV drugs is urgently required. Cyclophilin A (CypA) is a host factor required for HIV-1 replication, and plays a role in viral replication by interacting with the HIV-1 capsid protein (CA). As such, it represents a potential target for novel ARV drugs. We here searched for low molecular weight compounds that inhibited HIV-1 replication by interfering with binding between CypA and HIV-1 CA. A total of 106 compounds were screened in an in silico docking study as candidates that were predicted to interact with the HIV-1 CA binding pocket of CypA. Biological tests were then conducted to evaluate the anti-HIV-1 activities as well as cytotoxicities of these test compounds, and 4 compounds that efficiently inhibited viral replication without exhibiting strong cytotoxicity were subsequently selected. The molecular mechanisms underlying the inhibition of HIV-1 replication by the 4 selected compounds have not been elucidated in the present study; however, we consider that these compounds will become the lead compounds for developing novel ARV drugs once more detailed studies are performed on the molecular mechanisms responsible for their anti-HIV-1 activities.
| Abstract |
| Highly active antiretroviral (ARV) therapy has successfully reduced viral transmission, morbidity, and mortality associated with human immunodeficiency virus type 1 (HIV-1) disease; however, the emergence of drug-resistant viruses is a major obstacle associated with ARV therapy. Therefore, the development of a new class of ARV drugs is urgently required. Cyclophilin A (CypA) is a host factor required for HIV-1 replication, and plays a role in viral replication by interacting with the HIV-1 capsid protein (CA). As such, it represents a potential target for novel ARV drugs. We here searched for low molecular weight compounds that inhibited HIV-1 replication by interfering with binding between CypA and HIV-1 CA. A total of 106 compounds were screened in an in silico docking study as candidates that were predicted to interact with the HIV-1 CA binding pocket of CypA. Biological tests were then conducted to evaluate the anti-HIV-1 activities as well as cytotoxicities of these test compounds, and 4 compounds that efficiently inhibited viral replication without exhibiting strong cytotoxicity were subsequently selected. The molecular mechanisms underlying the inhibition of HIV-1 replication by the 4 selected compounds have not been elucidated in the present study; however, we consider that these compounds will become the lead compounds for developing novel ARV drugs once more detailed studies are performed on the molecular mechanisms responsible for their anti-HIV-1 activities. |
| Keywords |
| HIV-1; Low molecular weight compound; Cyclophilin A |
| Introduction |
| Highly active antiretroviral (ARV) therapy using two or more reverse transcriptase inhibitors and protease inhibitors for human immunodeficiency virus type 1 (HIV-1)-infected patients has achieved durable virological suppression and reduced HIV-1 transmission, morbidity, and mortality associated with HIV-1 disease. However, the emergence of drug-resistant viruses as a result of widespread drug use is currently recognized as one of the major obstacles associated with ARV therapy [1]. Therefore, a new class of ARV drugs that target other steps in the viral life cycle urgently needs to be developed in order to deal with multidrug-resistant viruses. In the present study, we searched for low molecular weight compounds that inhibited HIV-1 replication in order to accumulate information for the development of a new class of ARV drugs. |
| Cyclophilin A (CypA), a highly conserved peptidyl prolyl isomerase, was previously identified as the interacting cellular protein of the immunosuppressant drug, cyclosporin A (CSA) [2]. CypA was incorporated into HIV-1 particles by interacting with the CypA binding loop of the HIV-1 capsid protein (CA) during viral assembly in virus-producing cells [3-5]. The infectivity of HIV-1 particles was also found to be diminished by disruption of the CA-CypA interaction [6-9]. Subsequent studies revealed that CypA in virus-infected cells, but not in virus-producing cells played a major role in regulating viral replication [10,11], and the interaction between HIV-1 CA and CypA in virus-infected cells protected the virus from the inhibitory role of host restriction factor on viral replication [12]. Therefore, CypA is required for efficient HIV-1 replication, and, thus, is conceivable as a target for novel ARV drugs. |
| CypA has been shown to interact with HIV-1 CA through its hydrophobic, HIV-1 CA-binding pocket [13]. We herein selected 106 low molecular weight compounds that were predicted to interact with the pocket of CypA in an in silico docking study. Biological tests were then carried out to evaluate the anti-HIV-1 activities of the test compounds using a single round HIV-1 replication assay. |
| Materials and Methods |
| Cells |
| 293T cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco, Life Technologies, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) (Hyclone, Thermo Scientific, South Logan, Utah, USA). U87.CD4.CXCR4 (U87) cells were obtained from Drs. HongKui Deng and Dan R. Littman through the AIDS Research and Reference Reagent Program (ARRRP) (Division of AIDS, NIAID, NIH, USA), and were cultured in DMEM supplemented with 10% FBS, G418 (300 µg/ml), and Puromycin (1 µg/ml). MT4 cells were cultured in RPMI-1640 (Hyclone, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS. All cells were maintained at 37°C in a CO2 incubator. |
| Plasmids |
| The pNL4-3-derived, luciferase reporter proviral construct, pNL-Luc-E-R+ [14], and expression vector for vesicular stomatitis virus G glycoprotein (VSV-G), pHit/G [15], were used to generate VSV-G-pseudotyped HIV-1 (HIV-1/VSV-G). A proviral construct containing the CypA-binding-deficient CA, pNL-CA-G89A/P90A, was also constructed by site-directed mutagenesis using PfuUltra II Fusion HS DNA Polymerase (Agilent Technologies, Santa Clara, CA, USA) and the primers, 5’-GCATCCAGTGCATGCAGCGGCTATTGCACCAGG-3’ and 5’-CCTGGTGCAATAGCCGCTGCATGCACTGGATGC-3’ (mismatched nucleotides are underlined). Briefly, a 20 µl reaction mixture containing 0.2 µM of each primer, 500 µM of dNTP, 1 unit of PfuUltra II polymerase, and 4 ng of pNL-Luc-E-R+ was subjected to polymerase chain reaction (PCR). The PCR conditions used were as follows; enzyme activation at 95°C for 2 minutes; 24 cycles of 95°C for 10 seconds and 68°C for 8 minutes; and final extension at 68°C for 5 minutes. The PCR products were treated with Dpn I (New England Biolabs, Ipswich, MA, USA) to destroy the original templates before transforming E. coli HB101 competent cells with the PCR-generated mutant plasmid. The introduction of mutations was confirmed by sequencing. |
| Test compounds |
| One hundred and six commercially available compounds were selected in an in silico molecular docking study, as described previously [16]. This screening was performed by MOE (Chemical Computing Group). The crystallographic data of CypA in complex with Alanine-Proline dipeptide (PDB entry 2CYH) was obtained from the Brookhaven Protein Data Bank, and the structure of CypA was used as a receptor for screening. In the present study, the “MOE dock” program was used to perform all of the screening procedures. Triangle Matcher was used for placement, London dG was used for scoring function, MMFF94x forcefield was used for energy minimization after docking, and the other parameters were set as default. An active site covering the entire area of the 2 pockets in the CypA molecule was found using the SiteFinder module in MOE. We performed screening for alchemy database. Finally, candidates used for biological tests were selected according to the best docking score of each compound. Then, the compounds that were predicted to interact with the HIV-1 CA-binding pocket of CypA were designated IDs as CA1 to CA106. The test compounds were dissolved in dimethyl sulfoxide (DMSO) and diluted to a certain concentration with culture media to a final concentration of 0.2% DMSO. |
| Evaluation of antiviral activities of test compounds |
| The antiviral activities of the test compounds were evaluated by a single round HIV-1 replication assay using a luciferase reporter proviral construct. In this assay system, the late stage of HIV-1 replication, including the viral life-cycle steps, RNA transcription, protein translation, viral assembly, and the release of the progeny virus, could be monitored by evaluating the quantity and infectivity of the progeny virus released from proviral construct-transfected (virus-producing) 293T cells. In addition, the early stage of HIV-1 replication, including the viral life-cycle steps, viral entry into cells, encapsidation, reverse transcription, integration, RNA transcription, and protein translation, could be monitored by infecting HIV-1-permissive (target) cells with a reporter virus released from luciferase reporter proviral construct-transfected 293T cells, followed by measuring luciferase activity in infected cells. Briefly, 293T cells were seeded at 2 × 106 cells/10 ml in a 100 mm dish. Twenty-four hours later, the cells were co-transfected with pHit/G (1.18 µg) and the luciferase reporter proviral construct, pNL-Luc-E-R+ or pNL-CA-G89A/P90A (8.82 µg), using FuGENE HD transfection reagent (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Eighteen hours later, transfected 293T cells were trypsinized and re-seeded onto 96-well plates at a concentration of 2.8 × 104 cells/200 µl/well in the presence or absence of a testing compound. Forty-eight hours after being transfected, the cell culture supernatant of transfected 293T cells was collected. In the mode of action analysis, the virus quantity was evaluated by measuring the HIV-1 CA p24 antigen in samples using an ELISA kit (HIV-1 p24 Capture Assay, Advanced BioScience Laboratories, Rockville, MD, USA), according to the manufacturer’s instructions. Two HIV-1-permissive cell lines, MT4 and U87 cells, which were seeded on 96-well plates at concentrations of 2 × 104 cells/100 µl/well and 7 × 103 cells/100 µl/well 24 hours prior to the assay, respectively, were incubated with the culture supernatant of transfected 293T cells in the presence or absence of a test compound for 24 hours. Luciferase activities in virus-infected MT4 and U87 cells were then measured using the Steady-Glo Luciferase assay kit (Promega) with the microplate luminometer LB960 (Berthold, Bad Wildbad, Germany). |
| Cytotoxicity test |
| 293T, MT4, and U87 cells were seeded on 96-well plates at concentrations of 5 × 103 cells/100 µl/well, 2 × 104 cells/100 µl/well, and 7 × 103 cells/100 µl/well, respectively. After being incubated for 24 hours, cells were treated with a test compound in 200 µl of culture media. These cells were further incubated for 24 hours, and the proliferation of each cell line was measured using WST-1 reagents (Roche Diagnostics, Basel, Switzerland) according to the manufacturers’ instructions. |
| Analysis of virion-associated proteins |
| HIV-1/VSV-G carrying wild type or CypA-binding-deficient CA was produced by co-transfecting 293T cells with pHit/G and the proviral DNA, pNL-Luc-E-R+ or pNL-CA-G89A/P90A, respectively, as described above. A test compound was added 6 hours after transfection. Forty-eight hours after transfection, the culture supernatant of transfected 293T cells was collected and clarified by centrifugation at 6,000 x g for 10 minutes at 4°C. The virus in the culture supernatant was precipitated by ultracentrifugation through 20% sucrose at 140,000 x g for 2 hours at 4°C using Optima MAX-XP with a rotor, MLS-50 (Beckman Coulter, Brea, CA, USA). The viral pellet was resuspended in 140 µl of PBS. The virus quantity was evaluated by measured the HIV-1 CA p24 antigen in samples using an ELISA kit, as described above. Equivalent amounts of viral samples were loaded and separated on 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and then transferred onto PDVF membranes (Hybond-P, GE Healthcare Life Sciences, Uppsala, Sweden) using the Trans-Blot SD Semi-Dry Transfer Cell and PowerPac HC Power Supply (Bio-Rad , Hercules, CA, USA). Samples were then subjected to immunoblot analysis using either a rabbit anti-CypA polyclonal antibody (sc-20360-R; Santa Cruz Biotechnology, Dallas, TX, USA) or rabbit anti-HIV-1 p24 polyclonal antibody (65-004; BioAcademia, Osaka, Japan). After incubating samples with horseradish peroxidase-labeled, polyclonal goat anti-rabbit immunoglobulins (P0448, Dako, Troy, MI, USA), the antigen-antibody complex on the membrane was visualized using ECL prime (GE Healthcare Life Sciences), according to the manufacturer’s instructions. Virion-incorporated CypA and the HIV-1 CA p24 antigen were observed using the ChemiDoc XRS+ System (Bio-Rad). The expression of CypA in each sample was normalized to the expression of the HIV-1 CA p24 antigen. |
| Results |
| Screening for potent HIV-1 inhibitory compounds |
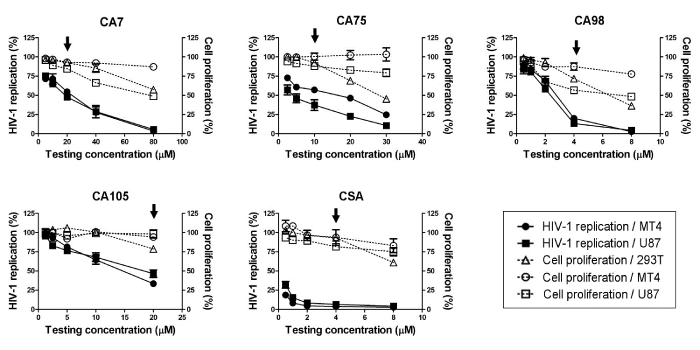
| In the first screening, all compounds were tested at a certain concentration according to their solubilities in DMSO, and to a concentration at which cells could survive. The compounds that inhibited HIV-1/VSV-G replication by more than 25% were considered to be the HIV-1 inhibitory compounds, while those that precipitated in culture condition were excluded. Although many of the 106 compounds tested in this study potently inhibited HIV-1, most also exhibited strong cytotoxicity, low solubility under the culture conditions, and/or did not exert inhibitory activity on viral replication in both the MT4 and U87 cell lines. Table 1 showed the properties of some compounds that could inhibit HIV-1 replication in the first screening. Four compounds: CA7, CA75, CA98, and CA105, which passed the screening criteria, were selected for further study. CA7, CA98, and CA105 exhibited strong HIV-1 inhibitory efficiencies as well as high solubilities in the culture condition. Although CA75 had a lower HIV-1 inhibitory efficiency than the 3 other compounds, it showed high solubility. Therefore, its antiviral effects were examined at higher concentrations, which were considered to be more effective. CA16, CA81, CA89, CA97, and CA106 may be able to inhibit HIV-1 replication more efficiently at higher concentrations. However, the tests for CA 16, CA81, and CA106 showed low reproducibility; and CA89 and CA97 had already been tested at a very high concentration. Therefore, these compounds were excluded from further studies. The antiviral activities and cytotoxicities of the 4 selected HIV-1 inhibitory compounds from the screening tests were further evaluated in a dose dependency manner. |
| Safety and Inhibitory Potency of the 4 selected compounds on viral replication in MT4 and U87 cell lines |
| The 4 selected compounds were evaluated for their antiviral activities and cytotoxicities in a dose dependency manner (Figure 1). CA7 inhibited HIV-1 replication in MT4 and U87 cells by 46% and 52%, respectively, at a concentration of 20 µM and the cell proliferation rates of 293T, MT4, and U87 cells in the presence of CA7 at that concentration were 92%, 93%, and 84%, respectively. CA75 inhibited HIV-1 replication in MT4 and U87 cells by 43% and 69%, respectively, at a concentration at 10 µM and the cell proliferation rates of 293T, MT4, and U87 cells in the presence of the compound at that concentration were 92%, 100%, and 88%, respectively. However, CA98 was markedly more toxic than the results from the screening indicated, and did not have a proper safe and potent concentration in the dose dependency evaluation. The most acceptable concentration was considered to be at 4 µM, a concentration at which HIV-1 replication was markedly decreased to 20% and 13% in MT4 and U87 cells, respectively, while the cell proliferation rates of 293T, MT4, and U87 cells in the presence of CA98 at that concentration were 72%, 87%, and 57%, respectively. CA105 inhibited HIV-1 replication in MT4 and U87 cells by 67% and 53% at a concentration at 20 µM, respectively, while the cell proliferation rates of 293T, MT4, and U87 cells in the presence of CA105 at that concentration were 78%, 95%, and 98%, respectively. CSA was included as a positive control in the experiment. It inhibited the HIV-1 replication in MT4 and U87 cells by 96% and 93%, respectively, and the cell proliferation rates of 293T, MT4, and U87 cells in the presence of CSA at that concentration were 93%, 94%, and 82%, respectively. Therefore, CA7, CA75, CA98, CA105, and CSA were subjected to further studies at concentrations of 20 µM, 10 µM, 4 µM, 20 µM, and 4 µM, respectively. |
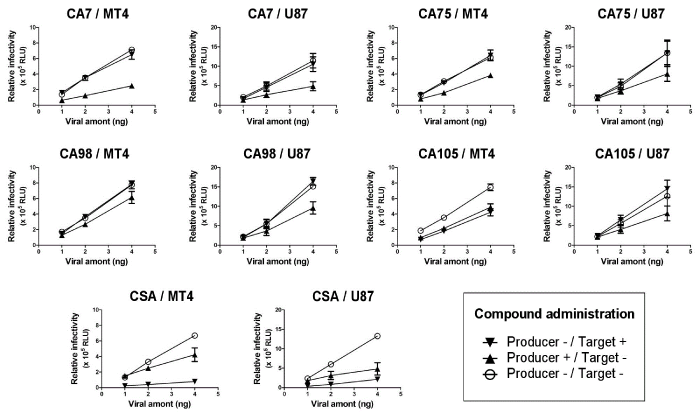
| Selected compounds inhibited HIV-1 replication in the virus-producing stage |
| In order to understand how the 4 selected compounds inhibited HIV-1 replication, their modes of action were investigated by observing the antiviral activity of each compound in virus-producing and -infected cells. Either virus-producing (proviral DNA transfected-) 293T cells or -infected MT4 or U87 cells were treated with the selected compound at the concentration indicated in Figure 2. The treatment of virus-producing 293T cells with the selected compounds reduced the infectivity of the progeny virus, although the level of virus production was not significantly affected by the compounds (data not shown). In contrast, the treatment of virus-infected MT4 or U87 cells with the compounds had no effect on viral replication, except for CA105, which also showed inhibitory effects on viral replication in MT4, but not in U87 cells. In contrast to the 4 selected compounds, CSA mainly inhibited viral replication by the treatment of virus-infected MT4 and U87 cells, and also exhibited viral inhibition by the treatment of virus-producing 293T cells. These results suggested that CA7, CA75, and CA98 had no inhibitory effects on the early viral replication step(s) in virus-infected cells, but inhibited HIV-1 replication presumably at the assembly step of viral particles in virus-producing cells. CA105 may also have an additional mechanism or use a different mechanism of inhibition than the 3 other selected compounds. These results suggested that the 4 selected compounds inhibited HIV-1 replication with modes of action related to viral production, but with a different mechanism to that of CSA. |
| Inhibitory effects of the 4 selected compounds may not have involved disruption of the CypA-CA interaction |
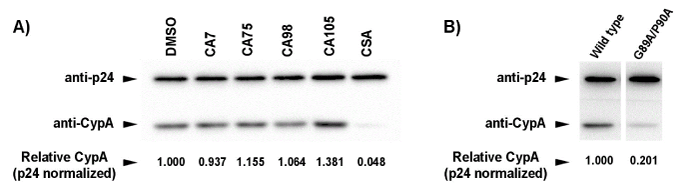
| Western Blot analysis was performed to evaluate whether the 4 selected compounds disrupted binding between human CypA and HIV-1 CA during the viral assembly process. CSA, as a positive control, markedly decreased the incorporation of CypA into the HIV-1/VSV-G virion. However, the incorporation rate of CypA into the HIV-1/VSV-G virion in the presence of CA7, CA75, or CA98 was similar to that in the negative control experiment including 0.2% DMSO, but was higher in the presence of CA105 (Figure 3A). These results indicated that the 4 selected compounds inhibited HIV-1 replication by an unknown mechanism other than disrupting the CypA-CA interaction, which is required for the incorporation of CypA into viral particles. |
| A CypA-binding mutant conferred resistance to CA75, but not to the 3 other compounds |
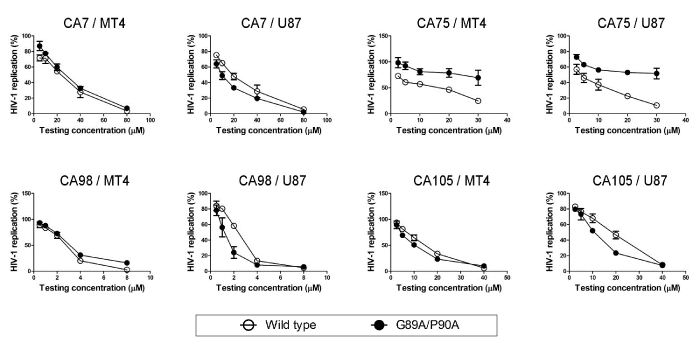
| To further verify that the 4 selected compounds inhibited HIV-1 replication by a mechanism other than disruption of the CypA-CA interaction, a HIV-1/VSV-G mutant containing CypA-binding-deficient CA was constructed and its replication was tested in the presence of the compounds. The introduction of the amino acid substitutions, G89A and P90A, into the CypA-binding loop of HIV-1 CA conferred a significant reduction in the incorporation of CypA into the HIV-1/VSV-G virion as expected (Figure 3B). The relative incorporation level of CypA was decreased to approximately 20% of the virus carrying the wild-type capsid (Figure 3B). The antiviral activities of the 4 selected compounds on HIV-1/VSV-G carrying wild type CA and the CypA-binding-deficient CA mutant were then evaluated in a dose dependency manner (Figure 4). The results obtained showed that the CypA-binding-deficient mutant could not confer efficient replication in the presence of CA7, CA98, and CA105 in both MT4 and U87 cell lines. In contrast, the mutation in the CypA-binding pocket could rescue 45% and 41% of viral replication in the presence of 30 µM of CA75 in both MT4 and U87 cell lines, respectively. Taken together with CypA being efficiently incorporated into viral particles in the presence of the 4 selected compounds (Figure 3A), these results suggested that CA7, CA98, and CA105 affected viral replication step(s) in virus-producing cells independent of the CypA-CA interaction. In addition, CA75 was suggested to inhibit viral replication by interfering with the CypA-CA interaction during virus assembly in virus-producing cells; however, it could not inhibit the incorporation of CypA into viral particles. |
| Discussion |
| There are countless numbers of low molecular weight compounds in the chemical databases of pharmaceutical companies. Molecular docking is an in silico technique that allows compounds with the desired functions to be selected. We aimed to select compounds that inhibited HIV-1 replication using a molecular docking study. However, molecular docking alone was not sufficient, and subsequent laboratory experiments were still required to confirm whether compounds was as effective in vitro as expected. In the screening tests, several compounds exhibited inhibitory effects on HIV-1 replication; however, some of these had strong cytotoxic effects while others exhibited low solubilities under the culture conditions used. If an antiviral compound could be found that efficiently inhibited HIV-1 replication with less toxic effects or no harm to human organs, patients may not develop adverse side effects. Cytotoxicity was generally measured as 50% cytotoxic concentration (CC50); however, it was impossible in the present study to evaluate CC50 because of the low solubilities of the test compounds. Four compounds: CA7, CA75, CA98, and CA105, were finally selected from the screening tests based on their inhibitory effects, cytotoxicities, solubilities, and reproducibilities. |
| Although the 4 selected compounds in the present study were predicted to bind to CypA in silico, no direct evidence was previous obtained for the binding of CypA in vitro. Western blot experiments revealed that none of the selected compounds inhibited the incorporation of CypA into the HIV-1 virion; however, they could inhibit HIV-1 replication in vitro. The infectivity of the progeny virus was reduced by treating virus-producing 293T cells with these compounds. These results suggested that the selected compounds inhibited HIV-1 replication by a mechanism other than that suppressing the incorporation of CypA into the virion. |
| The infectivity of the mutant virus with HIV-1 CA lacking CypA binding activity was reduced by CA7, CA98, and CA105, suggesting that these 3 compounds inhibited viral replication independent of CA-CypA binding. In contrast, the infectivity of the mutant virus was not inhibited by CA75, suggesting that CA75 blocked viral replication by interfering with the CA-CypA interaction in virus-producing cells. There is a discrepancy that CA75 might interfere the CA-CypA interaction, whereas it could not suppress the incorporation of CypA into virion. A possible explanation of the discrepancy is that CA75 might transiently interfere the binding of CypA to CA in virus-producing cells, and played a negative role on proper virus assembly; however, the stability or strength of CA75-CypA interaction might not be strong enough to inhibit the CypA-CA interaction throughout the virus assembly process. We consider that further studies are required to reveal the role of the compound in the process of virus assembly. In addition, it is necessary to study the possible incorporation of the compounds into viral particles. |
| Recent studies reported that CA-CypA binding in infected cells, but not in virus-producing cells played a critical role in the maintenance of viral infectivity, indicating that CypA in infected cells was more important than that in virus-producing cells [10-12]. In the present study, CSA inhibited viral replication both in virus-producing and -infected cells, while the 4 selected compounds inhibited viral replication in virus-producing cells (Figure 2). These results suggested that these compounds inhibited virus replication via a different mechanism to that of CSA. The mechanism by which the selected compounds inhibited HIV-1 replication has not yet been fully elucidated; however, further studies on the underlying mechanism(s) may contribute to revealing HIV-1 replication in more detail. Furthermore, the selected compounds are potentially lead compounds in the development of new antiretroviral drugs. Finally, we consider the limitation of the study is that we tested relatively small number of commercially available compounds on their antiviral effect. Many compounds selected by in silico study showed unfavorable characteristics, such as high cytotoxicity and low solubility, for testing by the subsequent in vitro study. It may be required to perform “back and forth” procedure between in silico and in vitro studies in order to optimize the screening process. In parallel with studying further the molecular mechanism(s) of how 4 selected compounds inhibited HIV-1 replication, it may be possible to perform more efficient screening tests to find potent and safe anti-HIV-1 compounds in a future study. |
| Conflict of Interest |
| The authors declare that there is no conflict of interest. |
| Acknowledgments |
| This work was supported in part by the program of the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) by the Ministry of Education, Culture, Sports, Science and Technology of Japan. U87.CD4.CXCR4 cells were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. The manuscript was proofread by Medical English Service (Kyoto, Japan). |
References
- Johnson VA, Calvez V, Gunthard HF, Paredes R, Pillay D, et al. (2013) Update of the drug resistance mutations in HIV-1: March 2013. Top Antivir Med 21: 6-14.
- Handschumacher RE, Harding MW, Rice J, Drugge RJ, Speicher DW (1984) Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science 226: 544-547.
- Franke EK, Yuan HE, Luban J (1994) Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372: 359-362.
- Luban J, Bossolt KL, Franke EK, Kalpana GV, Goff SP (1993) Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73: 1067-1078.
- Gamble TR, Vajdos FF, Yoo S, Worthylake DK, Houseweart M, et al. (1996) Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87: 1285-1294.
- Franke EK, Luban J (1996) Inhibition of HIV-1 replication by cyclosporine A or related compounds correlates with the ability to disrupt the Gag-cyclophilin A interaction. Virology 222: 279-282.
- Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh CT, et al. (1994) Functional association of cyclophilin A with HIV-1 virions. Nature 372: 363-365.
- Steinkasserer A, Harrison R, Billich A, Hammerschmid F, Werner G, et al. (1995) Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus type 1 (HIV-1): interference with early and late events in HIV-1 replication. J Virol 69: 814-824.
- Braaten D, Franke EK, Luban J (1996) Cyclophilin A is required for the replication of group M human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus SIV(CPZ)GAB but not group O HIV-1 or other primate immunodeficiency viruses. J Virol 70: 4220-4227.
- Sokolskaja E, Sayah DM, Luban J (2004) Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J Virol 78: 12800-12808.
- Hatziioannou T, Perez-Caballero D, Cowan S, Bieniasz PD (2005) Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. Journal of virology 79: 176-183.
- Towers GJ, Hatziioannou T, Cowan S, Goff SP, Luban J, et al. (2003) Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat Med 9: 1138-1143.
- Braaten D, Ansari H, Luban J (1997) The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J Virol 71: 2107-2113.
- Tokunaga K, Greenberg ML, Morse MA, Cumming RI, Lyerly HK, et al. (2001) Molecular basis for cell tropism of CXCR4-dependent human immunodeficiency virus type 1 isolates. J Virol 75: 6776-6785.
- Fouchier RA, Meyer BE, Simon JH, Fischer U, Albright AV, et al. (1998) Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J Virol 72: 6004-6013.
- Tian YS, Verathamjamras C, Kawashita N, Okamoto K, Yasunaga T, et al. (2013) Discovery of novel low-molecular-weight HIV-1 inhibitors interacting with cyclophilin A using in silico screening and biological evaluations. Journal of molecular modeling 19: 465-475.
Tables and Figures at a glance
| Table 1 |
Figures at a glance
 |
 |
 |
 |
|||
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
Relevant Topics
- Advanced Therapies
- Chicken Pox
- Ciprofloxacin
- Colon Infection
- Conjunctivitis
- Herpes Virus
- HIV and AIDS Research
- Human Papilloma Virus
- Infection
- Infection in Blood
- Infections Prevention
- Infectious Diseases in Children
- Influenza
- Liver Diseases
- Respiratory Tract Infections
- T Cell Lymphomatic Virus
- Treatment for Infectious Diseases
- Viral Encephalitis
- Yeast Infection
Recommended Journals
Article Tools
Article Usage
- Total views: 15387
- [From(publication date):
June-2015 - Jul 19, 2025] - Breakdown by view type
- HTML page views : 10693
- PDF downloads : 4694
