Screening Winter Wheat Lines in Nebraska for the Fhb1 Gene Using Kompetitive Allele Specific PCR (KASP)
Received: 23-Oct-2017 / Accepted Date: 28-Oct-2017 / Published Date: 15-Nov-2017
Abstract
Fusarium head blight (FHB) is one of the biotic stresses that causes a significant reduction in wheat grain yield and its quality. The main objective of this study was to screen a set of 53 high yielding Nebraska wheat lines (DUP15 - F6:7) for resistance to FHB using visual scoring and confirm the results using a Kompetitive Allele Specific PCR (KASP) marker for the Fhb1 gene. The experiment was conducted at Lincoln Field Station of the University of Nebraska-Lincoln. A high genetic variation was found between genotypes for their resistance to FHB. The phenotypic correlation S_FHB and grain yield was -0.61**. The results of the KASP assay revealed that only three genotypes NE15648, NE15674 and NE15569 had the predicted Fhb1 gene. KASP genotyping was a very effective technology which helped in screening valuable QTLs and associated trait markers. The three lines having the Fhb1 gene can be used in the wheat breeding program at University of the Nebraska-Lincoln to improve resistance to FHB.
Keywords: Wheat; FHB; KASP; Genetic variation; Triticum aesitvum L; Fusarium head bligh
Introduction
Fusarium head blight (FHB, caused by Fusarium spp .) or scab is a devastating disease of wheat (Triticum aestivum L. ). The disease causes a significant reduction in grain yield, seed quality and produces the mycotoxin deoxynivalenol (DON) that affects food safety and may cause serious health problems in human and domestic animals if the mycotoxin is present [1]. The disease continues to threaten susceptible wheat cultivars where environmental conditions such as high moisture and temperature persist during the flowering period. Significant grain losses due to FHB have been recorded in the history of wheat production in the U.S. FHB caused wheat losses at 2.72 million MT in 1982 and 4.78 million MT in 1993 [2]. The growing seasons in 2007, 2008, and 2015 had the highest FHB pressure in Nebraska which caused significant economic losses to wheat production. These losses indicated that winter wheat cultivars lack appropriate FHB resistance genes. The use of susceptible cultivars and rainfall during spring has contributed to the FHB epidemics in Nebraska.
Screening and identifying resistant genes in wheat germplasm for developing resistant varieties in combination with using cultural practices and fungicides are the most effective ways to manage FHB [3]. Resistance to FHB has been classified into multiple types. Type I resistance is attributed to reducing initial infection, type II resistance prevents spread of infection within the spike and type III confers resistance to mycotoxin accumulation [4]. Type II resistance (affected by Fhb1) has been widely used in breeding programs due to its effective performance in reducing FHB impact on grain production [5].
The advance in high-throughput genotyping methods has provided an explosion of single nucleotide polymorphism (SNP) sequences from plant genomes [6]. One of the advantages of these advances is the ability to convert the target SNPs into Kompetitive Allele Specific PCR (KASP) which is cost-effective, and introgression of target traits. Moreover, KASP is used in quantitative trait loci (QTL) mapping of parental populations, marker-assisted selection, and QTL fine mapping [7]. There are a lot of specific SSR, STS, AFLP, and RFLP markers that have been used to screen for the presence of the Fhb1 gene in wheat [8-12]. Although these markers are still highly used in molecular genetic studies, they are time consuming compared to the KASP technique. A wide range of species can be genotyped using KASP for various purposes (e.g. QTL mapping and GWAS). A KASP marker for the Fhb1 gene was used before for mapping the gene using QTL mapping [13].
The objectives of this study were to screen a segment of elite genotypes for the Fhb1 gene and ingrate those with Fhb1 gene in further breeding programs to improve resistance to FHB in a combination with high yield.
Material And Methods
Plant material and experimental layout
A set of 53 homozygous F3:7 lines from the preliminary yield trial (Nebraska Duplicate Nursery- DUP) were used in this study. Each line was derived from crosses between two different parents. Then, these lines were selected based on the grain yield, grain volume weight, and plant height [14]. The genotypes were sown on October 1st, 2014 in plots of 1.5 × 3 m with rows at 30 cm apart (four rows). The experiment was conducted at Lincoln Experimental Field Station of the University of Nebraska-Lincoln, USA.
Weather conditions
The 2015 growing season received continuous and above average rainfall during flowering period. The average flowering date was May 20th. The amount of precipitation in Lincoln research was 23.57 cm from May 1st to June 5th while normal precipitation is 11.18 cm during the reported period. Environmental conditions were quite conducive for FHB infection and development and 2015 was one of the worst growing seasons for FHB.
Phenotyping
The phenotyping or visual evaluation of FHB was done in the field 18 days after flowering. Susceptibility to FHB (S_FHB) was visually scored on 90 selected random heads per plot based on a scale of 1-10 where 1 is resistant and 10 susceptible [15].
DNA extraction and genotyping
DNA was extracted from the leaves of 53 genotypes using the BioSprint 96 automatic DNA extractor. The DNA concentration was diluted at 50 ng/μl in sterile distilled water to be used in KASP-SNP PCR reaction. All samples were arrayed in a 96 well plate. A 10 μl reaction with 5 μl DNA from each sample was mixed with 5 μl KASP reaction mix including a 0.14 μl of Fhb1 assay mix (LGC Genomics).
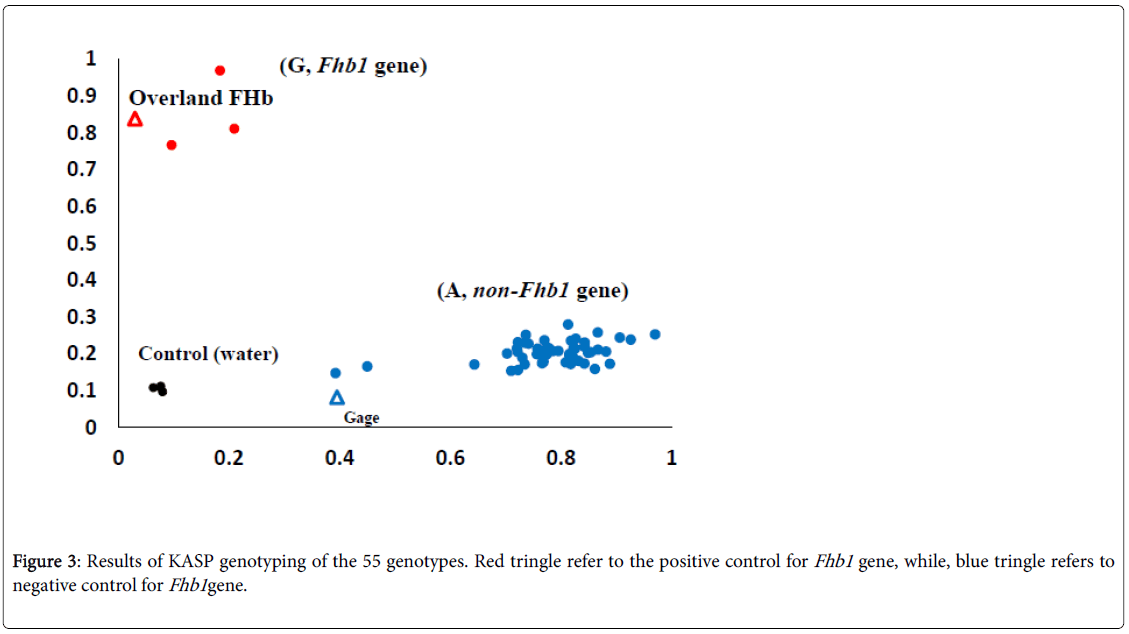
An Fhb1 KASP marker (wMAS000009) was ordered from LGCGenomics, Middlesex, UK. Thermal cycling conditions were 94°C for 15 min, followed by 10 cycles of touchdown PCR: 94°C for 20 s, 65– 57°C for 60 s (dropping 0.8°C per cycle), followed by 26 cycles of regular PCR: 94°C for 20 s, 55°C for 60s. The plate of samples was read by FLUO star Omega fluorescent. To determine the absence or presence of the Fhb1 gene, the allele G (presence) was labeled with HEX (red), on the other hand, the allele A (absence) was labeled with FAM (blue). To control the assay, two checks were used: Overland FHB1 (positive control for the Fhb1 gene) and Gage (negative control).
Statistical analysis
Single marker analysis was performed using R to test marker-trait association using the following model:
Y = μ + f (marker) + error
Where Y is equal to the trait value, μ is equal to the population mean, and f (marker) is a function of the molecular marker. All graphical presentations in this study were performed using R and Excel 2013.
Results And Discussion
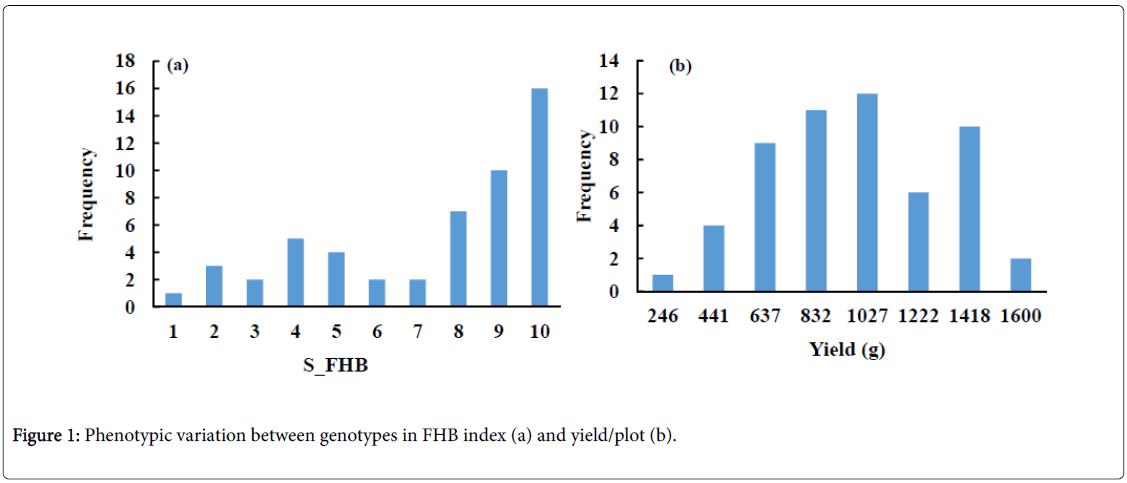
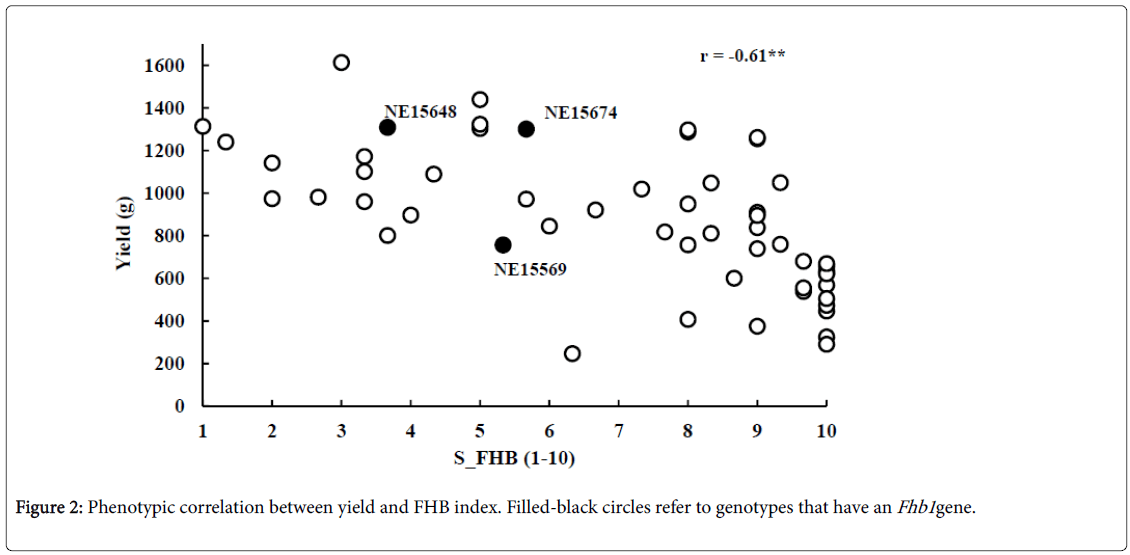
The phenotypic variation of FHB severity in the tested genotypes is illustrated in Figure 1a. The S_FHB ranged from 1 to 10 with an average of 7.08. Out of the 53 genotypes, 6 (11.3%), 11 (21.7%) and 37 (69%) were resistant (1-3) moderately resistant (4-6), and susceptible (7-9) to FHB, respectively. A high phenotypic variation was also observed in the yield of these genotypes (Figure 1b). The yield ranged from 249 to 1,613 g with an average of 879 g. A high negative significant correlation of r=-0. 61** was found between severity of FHB and grain yield (Figure 2).
The results of screening the 53 genotypes for the Fhb1 gene are presented in Figure 3 using KASP marker. The two alleles G and A are labelled red and blue in order to distinguish between genotypes in the current study. Out of the 53 genotypes, three (NE15674, NE15569, and NE15648) were found to carry the allele G which refers to the Fhb1 gene, while the rest of the genotypes carried the allele A (absence of Fhb1 gene).
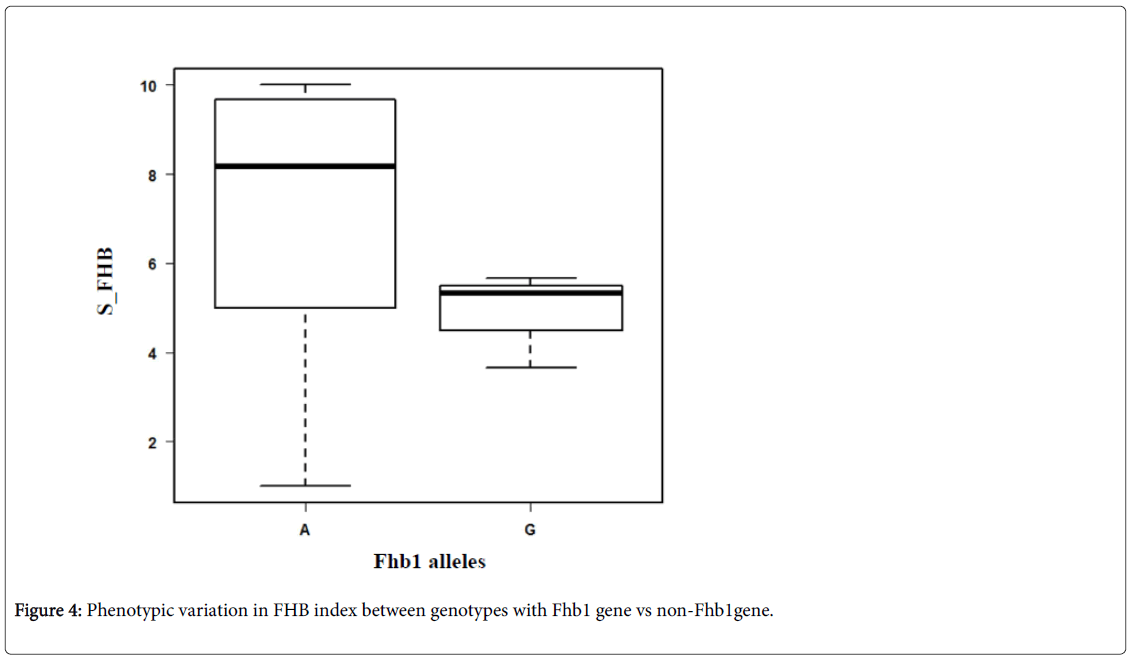
The variation between the two groups of genotypes (G vs. A) in their resistance to FHB is presented in Figure 4. On average, the genotypes which carry the Fhb1 gene (G) showed less symptoms (SI=4.89) than that do not carry the gene (A) (SI=8.3). However, six genotypes which carry allele A (non-Fhb1 gene) showed resistance to FHB with SI ranging from 1 to 3. Those genotypes probably carry one or more other genes for resistance to FHB (e.g. native resistance) Many earlier studies reported resistance QTL on 2B and 5AL chromosomes 3A [16], and 4AL and 6D3 [17]. All the markers used to detect the FHB genes are SSR, RFLP, STS, and AFLP which are less efficient compared to SNP markers. The KASP technology offers a costeffective, quick and efficient tool for introgression of target traits [18].
FHB resistance in wheat is complex and controlled by many genes in wheat [19,20]. The three genotypes which carry the Fhb1 gene showed higher yield with 757, 1,301 and 1,309 g for NE15569, NE15674, and NE15648, respectively. Different from other genes showing resistance to FHB, Fhb1 was verified effectively in other populations and utilized in many breeding programs to improve FHB resistance in wheat germplasm [21]. Fhb1 was first identified in Sumai-3 (variety included as a positive control). Fhb1 positive alleles occurred at a comparatively low level in all populations (2-10%).
Although the three genotypes carry Fhb1, they showed moderate resistance to FHB because Fhb1 is a quantitative resistance that confers partial and durable resistance. By looking at the pedigree of these lines, it seems that NE15674 (VA06W-558/TREGO FHB2//WESLEY FHB1) inherited Fhb1 gene from either TREGO FHB1 or WESLEY FHB1, NE15569 (NINGMAI 9415.16//SHA4/CHIL/3/NINGMAI 50/ NE01643//NE03458) inherited Fhb1 gene from NINGMAI, and NE15648 (NE91518 (Purple Wheat)/VA04W-474//1(ND2928/ Wesley//Wesley) F3/Wesley F3//HV9W03-539R)) inherited the gene from ND2928. On other hand, the wheat lines which were not confirmed to carry Fhb1 gene had parents with no known resistant gene in their pedigree. This result suggests the reliability of KASP markers in detecting the target genes in the tested genotypes. Moreover, it is very important to identify the source of the disease resistant genes for the forgoing assumption [22]. Although the three genotypes have Fhb1 gene, they exhibited different levels of FHB resistance (Figure 2). This result is most likely due to additional background genes affecting resistance. The researchers reported that Fhb1 generally reduced FHB based in the recurrent parent with no significant effect on yield and its quality [22].
Utilizing Fhb1 in wheat breeding program is very important for conferring durable resistance to FHB. Fhb1 confers type II resistance to FHB which prevents spread of the pathogen within wheat spike [23]. Therefore, the three lines having the Fhb1 gene can be used in breeding program in order to produce cultivars providing resistance to Fhb1. It seems that these three genotype show different level of moderate resistance to FHB. Therefore, crossing between these parents could be useful to develop resistant cultivars . For example, it was reported that a FHB resistant spring cultivar 'Sumai 3' was derived by crossing two moderately susceptible parents [24].
In conclusion, there are many challenges and difficulties to achieve reliable and reproducible FHB infection data due to the effects of the environment. Screening genotypes for FHB genes is the most effective way for improve FHB in wheat. The KASP technology provides reliable and accurate results to detect the target genes. This technology can be used for the marker-assisted selection by determining the characterization of genotype based on the genotypic level. The three wheat genotypes having Fhb1 gene can be integrated in breeding program to improve FHB resistant in winter wheat.
References
- Salgado JD, Madden LV, Paul PA (2015) Quantifying the Effects of Fusarium Head Blight on Grain Yield and Test Weight in Soft Red Winter Wheat. Phytopathology 105:295–306.
- McMullen M, Jones R, Gallenberg D (1997) Scab of Wheat and Barley: A Re-emerging Disease of Devastating Impact. Plant Dis 81:1340–1348.
- Wegulo SN, Baenziger PS, Hernandez Nopsa J, et al. (2015) Management of Fusarium head blight of wheat and barley. Crop Prot 73:100–107.
- David Miller J, Arnison PG (1986) Degradation of deoxynivalenol by suspension cultures of the fusarium head blight resistant wheat cultivar Frontana. Can J Plant Pathol 8:147–150.
- Cuthbert PA, Somers DJ, Thomas J, et al. (2006) Fine mapping Fhb1, a major gene controlling fusarium head blight resistance in bread wheat (Triticumaestivum L.). TheorAppl Genet 112:1465–1472.
- Semagn K, Babu R, Hearne S, Olsen M (2013) Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mol Breed 33:1–14.
- Gervais L, Dedryver F, Morlais JY, et al. Mapping of quantitative trait loci for field resistance to Fusarium head blight in an European winter wheat. TheorAppl Genet 106:961–970.
- Anderson JA, Stack RW, Liu S, Waldron L, Fjeld AD, et al.(2001) DNA markers for Fusarium head blight resistance QTLs in two wheat populations. TAG TheorAppl Genet 102:1164–1168.
- Otto CD, Kianian SF, Elias EM (2002) Genetic dissection of a major Fusarium head blight QTL in tetraploid wheat. Plant MolBiol 48:625–632.
- Zhou W, Kolb FL, Bai G (2002) Genetic analysis of scab resistance QTL in wheat with microsatellite and AFLP markers. Genome / Natl Res Counc Canada = Ge´nome / Cons NatlRech Canada 45:719–27.
- Guo PG, Bai GH, Shaner GE (2003) AFLP and STS tagging of a major QTL for Fusarium head blight resistance in wheat. TheorAppl Genet 106:1011–7.
- Holzapfel J, Voss HH, Miedaner T (2008a) Inheritance of resistance to Fusarium head blight in three European winter wheat populations. TheorAppl Genet 117:1119–28.
- Burt C, Steed A, Gosman N (2015) Mapping a Type 1 FHB resistance on chromosome 4AS of Triticummacha and deployment in combination with two Type 2 resistances. TheorAppl Genet 128:1725–38.
- El-basyoni I, Baenziger PS, Dweikat I (2013) Using DArT Markers to Monitor Genetic Diversity throughout Selection: A Case Study in Nebraska’s Winter Wheat Breeding Nurseries. Crop Sci 53:2363.
- RoelfsAP, Singh RP, Saari EE (1992) Rust Diseases of Wheat: Concepts and methods of disease management.
- Steiner B, Lemmens M, Griesser M (2004) Molecular mapping of resistance to Fusarium head blight in the spring wheat cultivar Frontana. TAG TheorAppl Genet 109:215–224.
- Paillard S, Schnurbusch T, Tiwari R (2004) QTL analysis of resistance to Fusarium head blight in Swiss winter wheat (Triticumaestivum L.). TheorAppl Genet 109:323–332.
- Curry JD, Darshna Y, Jon C (2016) Key Trait Screening on Global Wheat Accessions Using KASP Genotyping Markers - a New Open Resource for the Wheat Breeding Community.
- Holzapfel J, Voss HH, Miedaner T, Viktor Korzun, Jennifer Häberle, et al. (2008b) Inheritance of resistance to Fusarium head blight in three European winter wheat populations. TheorAppl Genet 117:1119–28.
- Buerstmayr H, Ban T, Anderson JA (2009) QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: a review. Plant Breed 128:1–26.
- Pumphrey MO, Bernardo R, Anderson JA (2007) Validating the QTL for Fusarium Head Blight Resistance in Near-Isogenic Wheat Lines Developed from Breeding Populations. Crop Sci 47:200.
- Bakhsh A, Mengistu N, Baenziger PS, et al. (2013) Effect of Fusarium Head Blight Resistance Gene on Agronomic and End-Use Quality Traits of Hard Red Winter Wheat. Crop Sci 53:793.
- Zhang J, Jin Y, Rudd J, Bockelman H (2008) New Fusarium head blight resistant spring wheat germplasm identified in the USDA National Small Grains Collection. Crop Sci 48:223–235.
Citation: Sallam A, Sidiqi J, Baenziger S (2017) Screening Winter Wheat Lines in Nebraska for the Fhb1 Gene Using Kompetitive Allele Specific PCR (KASP). J Plant Genet Breed 1: e104.
Copyright: ©2017 Sallam A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Open Access Journals
Article Usage
- Total views: 6140
- [From(publication date): 0-2017 - Dec 22, 2024]
- Breakdown by view type
- HTML page views: 5401
- PDF downloads: 739