Research Article Open Access
A New Enhanced, Rapid and Precise Sample Preparation Protocol for Label-free Protein Quantification
John R Griffiths1, Yvonne Connolly1, Ken Cook2, Kevin Meyer3 and Duncan L Smith1*
1Cancer Research UK Manchester Institute, University of Manchester, UK
2Thermo Fisher Scientific, Tudor Road, Manor Park, Runcorn, UK
3Perfinity Biosciences Inc., Indiana, USA
- *Corresponding Author:
- Duncan L Smith
Head of Mass Spectrometry
CRUKManchester Institute
Wilmslow Road, Manchester, M20 4BX, UK
Tel: +44-161- 446-8496
Fax: +44-161-918-7134
E-mail: duncan.smith@cruk.manchester.ac.uk
Received date: September 30, 2014; Accepted date: October 17 2014; Published date: October 21, 2014
Citation: Griffiths JR, Connolly Y, Cook K, Meyer K, Smith DL (2014) A New Enhanced, Rapid and Precise Sample Preparation Protocol for Label-free Protein Quantification. J Anal Bioanal Tech 5:213. doi: 10.4172/2155-9872.1000213
Copyright: © 2014 Griffiths JR, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
Label-free quantification using liquid chromatography-mass spectrometry (LC-MS) has now become a widely accepted analytical approach for the comparison of differential protein expression levels across multiple samples. One major concern of current label-free strategies is the technical variability introduced at multiple points during sample preparation. Typical workflows require cell lysis with buffers containing detergents, overnight proteolysis, removal of potential interferences such as salts and detergents by solid phase extraction (SPE) and subsequent solvent evaporation and reconstitution prior to analysis. Each of these stages is likely to introduce sample variability. Here we present a new strategy, which incorporates an acid-cleavable detergent in the lysis buffer, one-hour digestion with a temperature stable, immobilized enzyme and no requirement for SPE clean-up. The entire sample preparation stage takes less than three hours from cell pellet to autosampler vial and sample handling is kept to an absolute minimum. Our data demonstrate a significant reduction in the technical variability of sample preparation compared to a typical protocol along with a dramatic time saving with no cost in terms of qualitative peptide identifications.
Keywords
Proteomics; Protein quantification; Label-free; Immobilized trypsin
Introduction
Tandem mass spectrometry (MS/MS) in combination with nanoflow liquid chromatography (LC) has become the analytical technique of choice for global relative quantification assays of complex protein samples [1]. Methodologies have been developed to enable the comparison of protein expression levels across multiple samples, which may be broadly classified as either label-free, or those incorporating stable isotopes [2]. Stable isotopes may be introduced either at the point of cell growth (in vivo metabolic labeling) i.e. stable isotope labeling by amino acids in cell culture (SILAC) [3] or at the protein or peptide level via chemical derivatization (in vitro labeling with isotopomeric tags) such as reductive methylation [4], isobaric tags for relative and absolute quantification (iTRAQ) [5], tandem mass tags (TMT) [6] and isotopecoded affinity tags (ICAT) [7]. In contrast, label-free approaches take MS features of peptides such as peak intensity, or spectral numbers, and compare them directly across samples [8]. All of these approaches have advantages and limitations, which are briefly considered below.
Analytically speaking, when samples requiring quantitative comparisons of a biological nature are subjected to several stages of sample manipulation, it is advantageous to have an internal control (an isotopic label) present to compensate for variations introduced throughout the process i.e. to mitigate for technical variability. The earlier in the analytical process that this control can be incorporated, the less effect sample manipulation should have, since once the samples have been labeled they may be combined and subsequently experience all sample handling operations, such as enzymatic digestion, to the same extent. To this end, SILAC is the most attractive means of labeling samples for protein expression comparison. In a SILAC experiment, mass differences are introduced to all proteins by growing cells in normal and enriched media [3,9]. Normal media gives rise to a “light” cell population, whereas a “heavy” cell population is synthesized when this is replaced with media containing heavy amino acids, such as 13C6- arginine and/or 13C6-lysine. Upon incorporation of the heavy amino acids (five passages), the cells are mixed, digested and analyzed by LC-MS/MS, with quantitation achieved from MS peak intensities and identification from MS/MS spectra. The major limitations of a SILAC experiment are, firstly experimental cost, secondly metabolic labeling may not always is possible and finally the length of time required for analysis (five cell doublings for complete incorporation). SILAC experiments are also typically limited to the comparison of only three (light, intermediate and heavy) samples simultaneously.
means of chemically introducing isotopic labels at the protein level [7,10]. ICAT reagents comprise of three portions; a thiol-reactive group to enable protein attachment via cysteine residues, a linker region containing the stable isotopes and an acid-cleavable biotin group for subsequent affinity capture (avidin) enrichment. Original ICAT reagents consisted of light and heavy versions resulting from the incorporation of either eight deuteriums (heavy) or eight hydrogens (light) which gave rise to an 8 Da mass difference [7]. A subsequent version uses carbon-13 instead of deuterium to create the heavy form [10]. Samples are labeled, mixed, combined and digested before avidin affinity enrichment and mass spectrometric analysis. The ratio of MS peak intensities is a measure of the relative abundance of each peptide, and the MS/MS spectra enables protein identification to be inferred.
Labels may also be introduced chemically at a later point in the experiment, after cell lysis and usually post enzymatic digestion i.e. at the peptide level. Reductive methylation, also known as dimethyl labeling, is a cost effective, fast and efficient way to introduce stable isotopes to peptides [4,11]. By using combinations of light and heavy reagents (formaldehyde and sodium cyanoborohydride) primary amines (unmodified lysine residues and peptide N-termini) may be reductively methylated resulting in a peptide mass increase of 28.0313 Da (Light), 32.0564 Da (Intermediate) and 36.0757 Da (Heavy) per label introduced. After labeling, samples may be mixed (maximum triplex) and analyzed by LC-MS/MS with quantitation, similar to ICAT and SILAC, obtained from MS peak volumes. All of these strategies give rise to a more complex MS spectrum since for each peptide, up to three precursors, corresponding to the Light, Intermediate and Heavy isotope additions, are often present.
Unlike the approaches described above, which introduce mass differences as a result of the isotope introduction, isobaric tagging methods such as iTRAQ and TMT give rise to identical precursor masses with quantification obtained in MS/MS across up to eight channels [12,13]. Both of these types of reagent modify primary amines such as peptide N-termini and unmodified lysines with the isobaric tag. Upon CID fragmentation, reporter ions are released which contain different numbers of stable isotopes of carbon and nitrogen leading to ions at different m/z values. The intensity of these ions is proportional to the original amount of precursor ion from each labeled sample. Where multiple samples require comparison these types of reagent offer a clear advantage in terms of their multiplexing capabilities. However, they are relatively (compared to reductive methylation) expensive, require care in sample preparation to avoid interferences and are prone to an underestimation in fold differences [14]. In addition, quantitation is derived from MS/MS data, with only a few or single data points compared to label-free approaches which utilize multiple MS-based data points
In experiments that require comparison of only two samples, by far the simplest approach to quantification requires no isotopic labeling prior to analysis by mass spectrometry. Label-free quantification uses either precursor ion intensity [15] or spectral counting [16], to compare the relative abundance of samples. Since no isotopic labeling takes place, these are the least expensive and most straightforward approaches to relative quantification; however, they are also the most susceptible to technical variability introduced by sample handling. This is because any digestion, sample clean-up and evaporation steps are carried out independently and may introduce different levels of error. To improve the statistical power [17] of the analysis samples are typically analyzed by LC-MS/MS in replicate.
One major source of variability, and the rate-limiting step, associated with quantitative bottom-up proteomics is that of enzymatic proteolysis. Typically, protein mixtures in solution are digested with trypsin at 37°C for 12 hours or longer. Previous approaches used to reduce the digestion time include microwave-assisted digestion [18], the use of enzyme-friendly surfactants [19,20] and immobilized trypsin [21,22].
Here we present a new sample preparation strategy, using an acidlabile surfactant to aid cell lysis, and temperature stable immobilized trypsin capable of denaturing proteins and greatly improving enzyme kinetics. The new method is aimed at reducing both preparation time and the technical variability typically associated with label-free proteomics experiments and we assess its performance against a standard approach.
Experimental
Materials and chemicals
Temperature stable immobilized trypsin and digestion buffers for the Flash Digests were obtained from Perfinity Biosciences Inc. (West Lafayette, IN). Trypsin (sequencing grade), water (HPLC grade), formic acid (for mass spectrometry, ~98%), ammonium bicarbonate and trifluoroacetic acid (TFA, 99%) were obtained from Sigma-Aldrich Ltd (Poole, Dorset, UK). Benzonase nuclease (>99% purity) was obtained from Novagen, Merck chemicals (Darmstadt, Germany), HPLC grade acetonitrile was purchased from Fisher Scientific (Loughborough, Leicestershire, UK). RapiGest SF surfactant and SepPak C18 cartridges (100 mg) were obtained from Waters Corp. (Milford, MA).
Sample preparation
Cell pellets were lysed using ice-cold Perfinity Flash Digest buffer containing 0.1%w/w RapiGest surfactant and 2 μLmL-1 benzonase. Buffer (550 μL) was added to the cell pellet on ice, which was allowed to lyse for 30 minutes with vortexing every 10 minutes. Lysate was passed several times through a 23G gauge (0.6 mm) needle to sheer any remaining DNA rendering the sample suitable for accurate pipetting. 100 μL aliquots of lysate were removed for either Flash or overnight digestion. Typically, a suitable protein amount for Flash digestion will be in the range 1 ng to 1 mg (data not shown).
Flash Digests were carried out on an Eppendorf ThermoMixer C heated 96-well PCR plate shaker (Hamburg, Germany) at 70°C with constant agitation at 1400 rpm. After a suitable digestion time (optimized at 1 hour, see below), Rapigest was degraded by the addition of 400 μL of 0.1% TFA at 37°C for 40 min. Particulates were removed by centrifugation at 16,100 rcf for 10 minutes. Supernatant was transferred to an auto sampler vial for direct injection.
Overnight solution digests were carried out on an Eppendorf ThermoMixer Comfort heated shaker (Hamburg, Germany) at 37°C with agitation at 500 rpm. After digestion, Rapigest was removed as above, and 200 μL supernatant was transferred to an autosampler vial for direct injection. A further 200 μL was removed for clean-up. SPE clean-up procedures were carried out using a Waters 20-port vacuum manifold (Milford, MA) with Millipore vacuum pump (Billerica, MA) essentially as described in Villen and Gygi [23]. Briefly, peptides were bound to the C18 stationary phase in 0.1% TFA, washed to remove salts and detergents with further additions of 0.1% TFA, then eluted in 50:50 acetonitrile:water with 0.1% TFA added. After eluting from the C18 cartridges, peptides were dried and reconstituted into 200 μL of 0.1% TFA for direct injection onto the LC.
LC-MS/MS analysis
All LC-MS/MS analyses were performed on an LTQ Orbitrap XL mass spectrometer (Thermo Scientific, San Jose, CA) coupled to an Ultimate 3000 RSLC nano system (Thermo Scientific, formerly Dionex, The Netherlands). 1 μL injection volumes were used throughout and samples were loaded directly onto the analytical column, an Easy- Spray PepMap RSLC C18, 2 μm × 75 μm id × 50 cm (Thermo Scientific, formerly Dionex, and The Netherlands). The composition (v/v) of LC buffers were as follows; Buffer A - 99.9% water plus 0.1% formic acid and Buffer B - 80% acetonitrile, 19.9% water and 0.1% formic acid. Peptides were loaded directly onto the column at a flow rate of 400 nlmin-1 with an initial mobile phase composition of 1% B. The organic strength was increased linearly from 1% to 22.5% B over 17 minutes again at 400 nlmin-1, followed by an increase to 25.1% B over the next 2 minutes with a concomitant reduction in flow rate to 300 nlmin-1, and to 39% B over a further 11 minutes. A further increase to 60% B over the next 4 minutes was followed by a ramp to 95% B over 2 minutes where it was held for a further 2 minutes. The column was then allowed to re-equilibrate to 1% B for a total analysis time of 60 minutes.
The mass spectrometer was instructed to perform data dependent acquisition on the top six precursor ions, which were measured in the Orbitrap FTMS detector over the mass range 370-1200 m/z, at a nominal resolution of 60,000. MS/MS spectra were acquired in the ion trap under CID conditions with normalized collision energy of 35, isolation width of 3 Th, Q value of 0.25 and 30 ms activation time.
Data analysis
Xcalibur raw data files acquired on the LTQ-Orbitrap XL were directly imported into Progenesis LCMS software (Waters Corp., Milford, MA, formerly Non-linear dynamics, Newcastle upon Tyne, UK) for peak detection and alignment. Data were analysed using the Mascot [24] search engine using the parameter settings in Table 1.
| Enzyme | Trypsin |
|---|---|
| Number of missed cleavages | 1 |
| Database | Uniprot + TrEMBL combined |
| Taxonomy | Human |
| Variable Modifications | Deamidation (N, Q) Oxidation (M) |
| Peptide tolerance | +/- 15 ppm |
| MS/MS tolerance | 0.8 Da |
Table 1: Mascot searching parameters.
Results and Discussion
Optimal digestion time using flash digestion
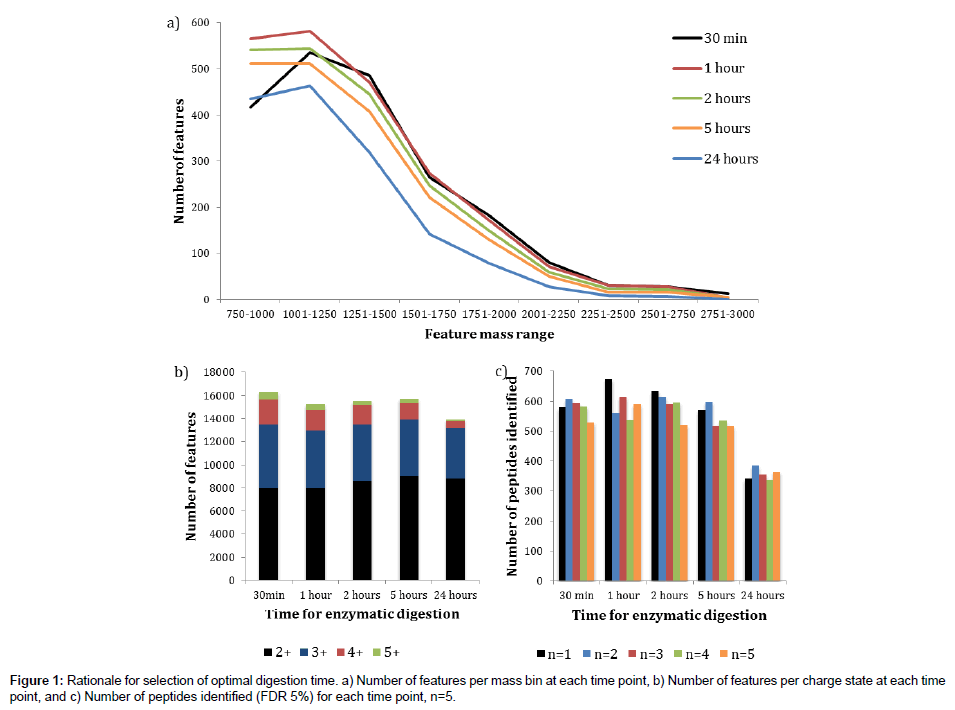
Prior to assessment of the new digestion protocol, it was first necessary to determine the optimal digestion time for the Flash Digest approach when applied to a complex mixture of proteins. Cell pellets were lysed as described in the Experimental Section above, then the lysates were simultaneously denatured and digested at 70°C with agitation for either 30 minutes, 1 hour, 2 hours, 5 hours or 24 hours.The digestion efficiency was assessed using post acquisition data processing with Progenesis LCMS software (Figure 1).
Figure 1a shows an overlaid plot of the number of features detected in Progenesis LCMS against feature mass range bins for each of the digestion times investigated. The trends show that the digestion times of 1 hour, 2 hours and 5 hours are very similar, with the 1 hour reaction time having more detected features than the other two for all of the selected feature mass ranges. A marked reduction in feature numbers is clearly seen after a 24 hours digestion period suggesting a level of deleterious degradation may be occurring. The 30 minutes digestion time plot follows the 1 hour plot very closely between the approximate mass range of 1500 Th and 2700 Th. However, there are far fewer features detected between 750 Th and 1250 Th and slightly more at the high mass end suggesting a moderate level of under digestion compared to the other time points. These data indicate that a 1 hour digestion time is optimal in terms of digestion efficiency of a complex protein mixture.
Next, using the same data set, we determined the number of features detected for a range of charge states (+2, +3, +4 and +5) at each digestion time (Figure 1b). There appears to be little difference observed for the 1 hour, 2 hours and 5 hours’ time points, with a slight increase in higher charges for the 30 minutes digest, and an overall reduction after 24 hours. Once again, there seems to be no justification for extending the digestion time beyond 1 hour. Finally, we assessed the data in terms of the number of peptides identified in a Mascot search (FDR 5%) with five replicate LC-MS/MS injections (Figure 1c). These data once again support the finding that extending the digestion time beyond 1 hour does not enhance the number of peptides identified, and that the number of identified peptides after a 24 hours digestion period is significantly reduced.
We conclude for these experiments that at a reaction temperature of 70°C, sufficient to denature cellular proteins whilst the immobilized trypsin remains fully active, a digestion time of 1 hour is optimal for a complex mixture of proteins such as that obtained from a whole cell lysate. All subsequent Flash Digest experiments presented in this paper were therefore performed with this digestion time.
Comparison of flash digest with overnight solution digestion ± SPE clean-up
Since the Flash Digest method offers significant advantages, in terms of speed of digestion, over existing approaches, we next wanted to assess the performance of this highly attractive protocol compared to a typical overnight solution digest both including and excluding a SPE clean-up step. Metrics used to assess the relative performance of the three approaches (Flash vs. Overnight vs. Overnight plus SPE) were i) number of peptides identified using a Mascot database search, and ii) technical variability introduced by each protocol.
When the five replicate injections for each of the three methods were combined into three concatenated files and analysed using the Mascot search engine, the number of peptides identified with both a 5% and 1% FDR were found to be very similar for the Flash Digest and overnight digest without SPE clean-up (Table 2). The data suggest that losses are incurred as a result of the clean-up stage where at 5% FDR, the number of peptides identified drops by approximately 14%. Taken together these data suggest that the Flash Digest performs as well as an overnight digest, and that the elimination of a clean-up step improves the peptide identification rate.
| FDR applied | Flash Digest | O/N digest | O/N digest + SPE |
|---|---|---|---|
| 5 % | 4335 | 4306 | 3710 |
| 1 % | 2507 | 2362 | 2146 |
Table 2: Number of peptide identifications using a Mascot search.
In addition to peptide identification, arguably of more importance to a label-free experiment is the amount of technical variability within an experiment. Since the principle aim of such an experiment is to detect true biological differences between samples, the sensitivity of the method is greatly improved by reducing non-biological differences i.e. technical variance or noise. We postulated that the amount of technical variability introduced at the sample preparation stage of a label-free experiment would be significantly lower if the samples were subjected to less manual handling procedures, such as SPE clean-up and evaporation to dryness. In addition, the use of immobilized trypsin negates the requirement of a protein assay being carried out since the enzyme:substrate ratio is less critical.
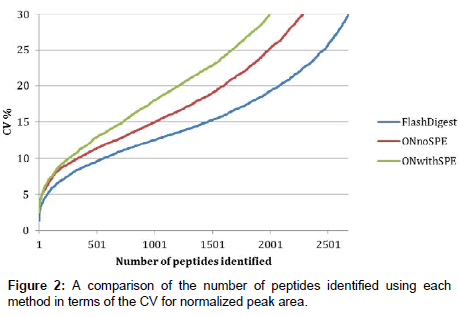
Figure 2 shows a comparison of the technical variability of the three methods investigated. Each method was performed in quintuplicate and each of these samples was injected as five replicates onto the LC-MS. In order to compare the overall technical variability associated with each analytical method, the normalized peak areas, taken from Progenesis LCMS, for all identified peptides with a Mascot ion score cut-off of 20 were analyzed in an Excel workbook as follows. Firstly, each digest (n=5) had five (n=5) replicate injections and a peak area associated for each. It was therefore possible to calculate a mean peak area for each identified peptide (present in all five replicate injections). These mean values were next compared across the five digests to obtain a % CV value for each peptide. Figure 2 represents these % CV values plotted against the number of identified peptides with such a % CV for each of the methods tested. By reading across from any chosen % CV value on the y-axis, it is possible to compare the number of peptides, for example at 15% CV, there are approximately 1500 peptides identified using the Flash Digest method, whereas only 1000 and 700 are identified in the overnight digest and overnight digest plus SPE methods respectively.
We can therefore conclude from this that the Flash Digest method is more precise than the other two, and that the technical variability associated with this approach is significantly lower. The data clearly show that the Flash Digest protocol results in more reproducible quantitation measurements than overnight digestion and that SPE does indeed compound the issue further. The main consequence of this reduction in variance is an increase in the sensitivity of the method to detecting biological differences between samples.
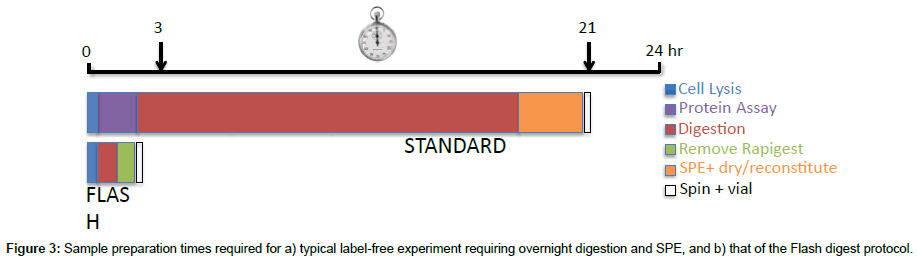
The final factor to consider in the assessment of the Flash digest protocol is the significant reduction in time required to process a cell pellet for LC-MS/MS analysis. The total sample preparation time from cell pellet to sample vial using the Flash Digest method is less than 3 hours. This is primarily a result of the extremely fast digest kinetics, which negate the need for an overnight digestion. Other time savings are associated with the method not requiring a protein assay to be performed, or for a sample clean-up step prior to direct on-column loading (Figure 3).
Conclusions
When compared to a commonly adopted label-free workflow used in proteomics, our new, simplified, reproducible method offers a number of distinct advantages. Firstly, the method described is simple, and may be easily adopted by non-specialist laboratories with little training or experience of proteolysis. Secondly, samples are lysed, digested and introduced to the mass spectrometer within a three-hour time frame without the need for detergents, chaotropes or reduction/ alkylation steps. This rapid preparation time is vastly superior to those requiring overnight digestion and helps to ensure sample integrity due to the decreased opportunity for sample degradation and undesirable modification. Thirdly, digests are shown to be more reproducible using the temperature stable immobilized trypsin of a Flash Digest compared to standard solution digests. Accompanying this is a dramatically reduced sensitivity to enzyme: substrate ratio, which for the purposes of these assays negates the need for a protein assay to be performed prior to digestion. Fourthly, as a consequence of the use of LC compatible buffers and an acid-cleavable detergent for cell lysis, no sample cleanup is required prior to LC-MS/MS even with direct injection nanoflow LC. Finally, as a result of the points described above, the new method is shown to be more analytically robust with tighter CV values on peptide peak areas of peptides identified which should enable more subtle differences in protein expression to be detected between samples.
Conflict of Interest Disclosure
K.M is Director of Research and Development at Perfinity Biosciences who manufacture the Flash Digest kit. The remaining authors declare no competing financial interest.
Acknowledgements
J.G, Y.C and D.S were funded by Cancer Research UK. We thank Dr Mark Holland (formerly of CRUK Manchester Institute) for providing the cell pellets.
References
- Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem 389: 1017-1031.
- Rodríguez-Suárez E, Whetton AD (2013) The application of quantification techniques in proteomics for biomedical research. Mass Spectrom Rev 32: 1-26.
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, et al. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1: 376-386.
- Hsu JL, Huang SY, Chow NH, Chen SH (2003) Stable-isotope dimethyl labeling for quantitative proteomics. Anal Chem 75: 6843-6852.
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, et al. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3: 1154-1169.
- Dayon L, Hainard A, Licker V, Turck N, Kuhn K, et al. (2008) Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal Chem 80: 2921-2931.
- Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, et al. (1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol 17: 994-999.
- Zhu W, Smith JW, Huang CM (2010) Mass spectrometry-based label-free quantitative proteomics. J Biomed Biotechnol 2010: 840518.
- Ong SE, Mann M (2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat Protoc 1: 2650-2660.
- Shiio Y, Aebersold R (2006) Quantitative proteome analysis using isotope-coded affinity tags and mass spectrometry. Nat Protoc 1: 139-145.
- Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc 4: 484-494.
- Unwin RD, Griffiths JR, Whetton AD (2010) Simultaneous analysis of relative protein expression levels across multiple samples using iTRAQ isobaric tags with 2D nano LC-MS/MS. Nat Protoc 5: 1574-1582.
- McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, et al. (2012) Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal Chem 84: 7469-7478.
- Ow SY, Salim M, Noirel J, Evans C, Rehman I, et al. (2009) iTRAQ underestimation in simple and complex mixtures: "the good, the bad and the ugly". J Proteome Res 8: 5347-5355.
- Bondarenko PV, Chelius D, Shaler TA (2002) Identification and relative quantitation of protein mixtures by enzymatic digestion followed by capillary reversed-phase liquid chromatography-tandem mass spectrometry. Anal Chem 74: 4741-4749.
- Lundgren DH, Hwang SI, Wu L, Han DK (2010) Role of spectral counting in quantitative proteomics. Expert Rev Proteomics 7: 39-53.
- Levin Y (2011) The role of statistical power analysis in quantitative proteomics. Proteomics 11: 2565-2567.
- Lill JR (2009) Microwave Assisted Proteomics. Royal Society of Chemistry.
- Ross AR, Lee PJ, Smith DL, Langridge JI, Whetton AD, et al. (2002) Identification of proteins from two-dimensional polyacrylamide gels using a novel acid-labile surfactant. Proteomics 2: 928-936.
- Yu YQ, Gilar M, Lee PJ, Bouvier ES, Gebler JC (2003) Enzyme-friendly, mass spectrometry-compatible surfactant for in-solution enzymatic digestion of proteins. Anal Chem 75: 6023-6028.
- Ma J, Lui J, Sun L, Gao L, Liang Z, et al. (2009) Online integration of multiple sample pretreatment steps involving denaturation, reduction, and digestion with microflow reversed-phase liquid chromatography-electrospray ionization tandem mass spectrometry for high-throughput proteome profiling. Anal Chem 81: 6534-6540.
- Sun L, Zhu G, Yan X, Mou S, Dovichi NJ (2014) Uncovering immobilized trypsin digestion features from large-scale proteome data generated by high-resolution mass spectrometry. J Chromatogr A 1337: 40-47.
- Villén J, Gygi SP (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc 3: 1630-1638.
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551-3567.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 14963
- [From(publication date):
November-2014 - Nov 21, 2024] - Breakdown by view type
- HTML page views : 10488
- PDF downloads : 4475