Saccharification and Bioethanol Fermentation of Carbohydrate-Extracted Microalgal Biomass by Genetically Identified Organisms
Received: 04-Dec-2017 / Accepted Date: 18-Jan-2018 / Published Date: 25-Jan-2018 DOI: 10.4172/2155-952X.1000279
Abstract
Saccharification of biomass to fermentable sugar is a major constraint for bioethanol production due to high cost of enzyme production and complications associated with the removal of hearse acid, alkali and salts formed after neutralization. This led to the search for low cost enzyme and its combination with dilute acid to enhance biomass hydrolysis. In this study, the microalgal biomass was hydrolysed using amylase and cellulase enzymes produced by solid state and submerged fermentation processes. Saccharification of algal biomass was studied using dilute tetraoxosulphate (VI) acid, crude enzyme complex and a combination of both. The highest yield of reducing sugar of 0.63 mg/ml was obtained with the co-combination hydrolysis of acid and enzyme, followed by acid hydrolysis (0.41 mg/ml) while the least was found with enzyme hydrolysis (0.36 mg/ml). The hydrolysate of the algal pretreated biomass was used for bioethanol production by Saccharomyces cerevisiae and co-cultures of S. cerevisiae and Aspergillus niger. The highest ethanol yield of 0.33 mg/ml at a percentage of 10.82% v/v was obtained from hydrolysates pretreated with co-combination of dilute acid and crude enzyme complex. The result showed that crude enzyme can increase the yield of hydrolyzed microalgal biomass for bioethanol production.
Keywords: Crude enzyme; Fermentation; Hydrolysis; Microalgae; Saccharification
Introduction
The world growing interest on alternative energy sources has attracted researcher’s interests to focus on biomass as a cost effective and efficient feedstock for bioethanol production and can serve as a suitable transportation fuel replacing the limited crude oil [1]. They have fast growth rate, higher photosynthetic efficiency, ability to fix greenhouse effects, non-competitive nature with food production, easily cultivated in non-agricultural site, high carbohydrate content and contain cellulose without lignin [2,3].
The microalgal biomass contains high carbohydrate from cellulose and hemicelluloses on its cell walls and starch with in the chloroplasts. Several microalgae species have the ability to generate elevated level of carbohydrate rather than lipid as stored polymers. The heterogeneous composition of the biomass, intracellular starch granules and cellulytic cell wall requires the exploitation of high cost available cocktail of carbohydrases to ensure total hydrolysis [4]. Successful utilization of algal biomass as bioethanol feedstock is dependent on development of economically feasible process technologies to ensure total hydrolysis of the biomass to simple low molecular weight sugar. Such technologies like solid state fermentation involving low cost biomass residues for enzymes production by suitable microorganism can be easily utilized for the hydrolysis in bioethanol fermentation process. Pretreatment process increases the crystalline nature of cellulose while enabling enzymatic degradation. Dilute acid pretreatment can increase the surface area of the carbohydrate availability thereby enhancing its reactivity with the enzymes and its biotransformation [5]. Microalgal biomass contains starch, cellulose and lipid combined with in its complex structure. The starch is commonly degraded by these enzymes, amylases and cellulase by cellulose, produced by several microorganisms such as bacteria and fungi. The majority of the commercial and laboratory cellulases are synthesized by fungi due to their high enzyme activities but several factors suggest that bacteria may have better excellent potential [6]. Bacteria frequently have higher growth rates than fungi allowing for higher rate of enzyme production. Most significantly they show affinity for heat stablility and are easier for genetic purpose. Amylase production using synthetic media is expensive and this stimulates search for an inexpensive route for its production by researchers. Solid state fermentation has been reported as a cost effective method to be employed in enzyme production and very vital in amylase production [7]. Agro industrial wastes have been reported to be a good substrate for the cost effective production of alpha amylase and thus have attracted many researchers [8,9].
In this study, solid state and submerged fermentation processes was used on the saccharification of microalgal biomass from plantain peel using dilute tetraoxosullphate (VI) acid, enzymes and co-combination of both. The bioethanol fermentation of the hydrolyzed microalgal biomass was examined using Saccharomyces cerevisiae and co-culture of A. niger and S. cerevisiae, respectively.
Materials and Methods
Sample collection, extraction and blooming
Unripe plantain peels were collected from different market sources in Port Harcourt, washed, sun dried and grinded using a manual hand grinding machine (Corna model 562). This was kept for further use in a sterile bottle. A novel synthetic medium [10] containing antibiotics (tetracycline and Nystatin to eliminate bacteria and fungi growth, respectively), was used to isolate Chlorella species. Agar plate technique was used for the isolation of the cultured plates and incubated for 3-5 days in a shade under natural illumination (sunlight).
Microorganisms screening and isolation
Bacteria were isolated from the soil using spread plate technique on carboxyl methyl cellulose (CMC) agar media. The plates were incubated at 45°C for 24 h, flooded with 0.1% aqueous solution of Congo red and rinsed with 1 M NaCl to detect the hydrolyzed zones [12]. To screen for the cellulase activities of the organisms, the diameter of the clear zones around the colonies on CMC agar were measured. A bacterial isolate with the highest diameter was selected for cellulase production.
Fungi was isolated from spoilt food samples using spread plate technique on PDA agar media containing lactic acid to prevent bacterial contamination. The plates were incubated at 30°C for 3 days. To visualize the hydrolysis zone, the plates were flooded with iodine and observed after 10 min. To indicate the amylase activities, the diameters of the clearance zones were measured and the isolate with the highest diameter was selected for amylase production. The organisms were identified using molecular and phylogenetic analysis method [13-15].
Production, extraction and enzyme assay
Cellulase production media containing the following in g/l; cellulose substrate (CMC), 10; MgSO4.7H2O, 0.3; K2HPO4, 2.0; (NH4)2SO4, 2.5; peptone, 10; were inoculated with the 24 h old bacterial culture and incubated at 37°C in a shaker incubator for 24 h. After fermentation, the culture media were centrifuged at 5000 rpm for 15 min to obtain the crude extracts which serve as the crude enzyme. Cellulase activity of the crude extract was assayed following the method of Miller [16]. 0.2 ml of the extract was added to 1.8 ml of 0.5 mM sodium phosphate (pH 7) and incubated in a water bath at 37°C for 30 min. The reaction was terminated by adding 3 ml of DNS reagent and boiled for 5 min and absorbance was measured at 575 nm.
Amylase production was assayed following the method by Singh et al. [17]. Powdered plantain peel substrate was mixed with mineral medium containing the following salts in g/l; soluble starch, 5.0; yeast extracts, 2.0; KH2PO4, 1.0; MgSO4.7H2O, 0.5; to moist the substrate. The mixture was inoculated with 3 day old fungi isolate dislodged with 0.1% tween 80 and incubated at 30°C for 5 days. After fermentation, the product was recovered by adding 0.1 M citrate buffer vigorously shaken for 30 min. The mixture was filtered and centrifuged at 5000 rpm for 20 min. The supernatant was filtered to obtain cell free extract which serves as crude enzyme source [18]. Amylase activity of the crude extract was estimated following method adopted by Miller [19]. About 1 ml of the crude extract was mixed with 1 ml of 1% soluble starch in 0.1 M citrate buffer (pH 5.0) and incubated in a water bath at 45°C for 30 min. The enzyme activity was terminated by adding 3 ml DNS reagent and boiled for 15 min and the absorbance measured at 575 nm.
Microalgal biomass hydrolysis
The micro algal biomass obtained was sectioned into three:
The first section was subjected to acid hydrolysis following method by Miranda et al. [20]. The algal biomass was hydrolyzed using 10% of 2 N H2SO4, autoclaved at 121°C for 45 min. The solution obtained was neutralized with 4 N NaOH, centrifuged at 7000 rpm for 20 min and filtered.
The second section was subjected to enzyme hydrolysis adopting method by Dhull et al. [21]. The algal biomass was incubated with 3% each of crude amylase and cellulase preparations obtained from enzyme production. The mixture was maintained at pH 4.8 and incubated at 50°C for 24 h. Then the mixture was centrifuged at 9000 rpm for 15 min and filtered.
The third section was subjected to combined acid and enzyme hydrolysis. The algal biomass was hydrolyzed with 10% 2 N H2SO4, at 121°C for 45 min and then neutralized to pH 4.8 with citrate buffer. This was incubated with 3% each of crude amylase and cellulase enzyme preparations and kept in a water bath for 12 h. Then centrifuge at 9000 rpm for 15 min and filtered to obtain clear supernatant.
These three sections of hydrolysates were analyzed for total reducing sugars adopting the modified procedure of Dubois [22].
Fermentation of microalgal hydrolysates
Saccharomyces cerevisiae was isolated from fresh palm wine and used for ethanol fermentation. A 12 h old S. cerevisiae culture cultivated in a Yeast Extract Potato Dextrose (YEPD) broth and a 3 day old A. niger culture in Potato Dextrose Broth were used to carry out the fermentation process for 5 days at 30°C.
The hydrolysates of the microalgal biomass obtained were supplemented with the following nutrient in g/l; Ammonium sulphate, 2; potassium monophosphate, 1; potassium dihydrogen phosphate, 1; zinc sulphate, 0.2; magnesium sulphate, 0.2 and yeast extract, 2. The media were sterilized at 121°C for 25 min and pH adjusted to 4.5 [23]. They were subjected to fermentation by S. cerevisiae and co-culture fermentation by S. cerevisiae and A. niger.
Analytical Procedures
About 5 ml of the samples were collected at intervals daily, centrifuged at 4000 rpm for 30 min to remove the cells from the supernatant and filtered for ethanol and reducing sugar determination [24].
Reducing sugar determination
Reducing sugar content was estimated using DNS method [19]. About 1 ml of the cell sample was mixed with 2 ml of the DNS reagent, placed in boiling water bath for 10 min and 7 ml of distilled water added. The absorbance was measured at 540 nm using UV Spectrophotometer (Model 721) [25].
Ethanol determination
The method of Caputi et al. [26] was used for ethanol content estimation. About 1ml of the cell sample was added to 1ml of chromic reagent and incubated in a water bath at 80°C for 10 min. They were then allowed to cool and absorbance analyzed at 600 nm. Ethanol quantity was extrapolated from standard ethanol curve.
Distillation
After 5 days of fermentation, the resulting cultured broth was centrifuged at 7000 rpm for 30 min, distilled with fractional distillation unit at 75°C [27]. The percentage of the ethanol in the distillate was estimated from ethanol standard curve by determining the ethanol concentration of the distillate and extrapolating them from series of standard ethanol/water mixture [28, 29].
Results
Attributes of Cellulase Producing Bacteria and Amylase Producing Fungi out of eleven bacterial isolates from the soil sample and spoilt food materials from the refuse dump site, only six showed halo zones of clearance around the growing colonies. They were selected as cellulase producing bacteria (Table 1).
| Isolate code | Zones of clearance in mm |
|---|---|
| BS1 | NT |
| B29 | 4.2 |
| BS3 | NT |
| BS4 | 2 |
| BS5 | NT |
| B30 | 3.8 |
| B31 | 4 |
| BY8 | 2.2 |
| BY9 | NT |
| BO10 | 3.2 |
| BO11 | NT |
Table 1: Sizes of clearance zones of bacterial isolates.
The three different moulds isolated from the spoilt food sample had colonies appearing cracked behind PDA plates and showing zones of clearance around the mycelia growth on starch media by addition of iodine indicating Aspergillus sp. The isolate showing the widest zone of clearance was selected as amylase producing fungi.
Molecular identification of the isolates
The results of genomic DNA quantification of the isolates using NanodropTM spectrophotometer and agarose gel electrophoresis showed that the DNAs extracted from the isolates were pure (Table 2). The plates showed the 16S rRNA gene PCR amplification bands of the bacteria isolates and 18S rRNA gene PCR amplification bands of the Fungi isolates applied in the study (Plates 1 and 2) (Figures 1 and 2).
| Samples | DNA conc. (ng/uL) |
Mean Absorbance (nm) | ||
|---|---|---|---|---|
| 260 | 280 | 260/280 | ||
| BS2 | 46.45 | 0.929 | 0.463 | 2.01 |
| BS2 | 46.45 | 0.314 | 0.148 | 2.12 |
| BS6 | 15.68 | 0.283 | 0.122 | 2.33 |
| FM1 | 14.16 | 0.283 | 0.122 | 2.33 |
| FY12 | 6.62 | 0.132 | 0.063 | 2.09 |
| BY7 | 6.49 | 0.13 | 0.052 | 2.49 |
| FY1 | 3.77 | 0.075 | 0.022 | 3.37 |
| A1 | 6.64 | 0.133 | 0.057 | 2.31 |
Table 2: Genomic and quantification using nano drop spectrophotometer.
The cellulase activities of the three selected bacteria isolates in cellulase production media showed that B30 had the highest cellulase activity of 1.48 μ/ml while the B29 and B31 had the lowest cellulose activities of 1.26 and 1.32 μ/ml (Table 2). B30 was therefore selected for enzymatic hydrolysis of the microalgae biomass. The Amylase activity of the two selected Aspergillus Isolates in amylase production media showed that FM1 had higher amylase activity of 1.52 μ/ml and therefore selected for cellulose hydrolysis of the microalgae (Figure 3).
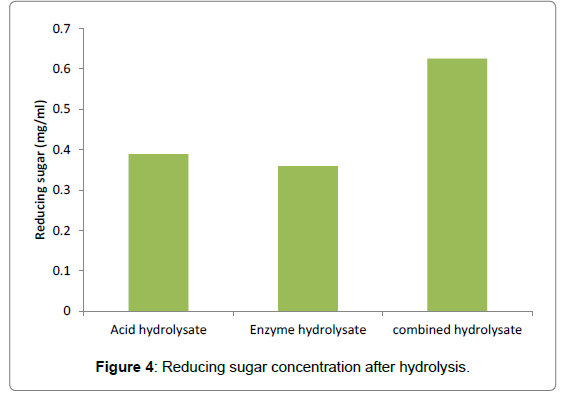
Three different saccharification treatments were employed for hydrolysis of the algal biomass. Dilute acid hydrolysis was performed with 2 N tetraoxosulphate (VI) acid, enzyme hydrolysis using enzyme complexes of amylase and cellulase and a combination of acid and enzyme hydrolysis. Test for reducing sugars in the hydrolysates showed that combined acid and enzyme hydrolysis produced the highest amount of reducing sugars of 0.63 mg/ml while acid and enzyme hydrolysates gave the lowest amount of reducing sugars of 0.39 mg/ml and 0.36 mg/ml, respectively (Figure 4).
The microalgal biomass hydrolysates obtained were subjected to fermentation by S. cerevisiae and co-culture fermentation by S. cerevisiae and A. niger.
Reducing sugars during the fermentation
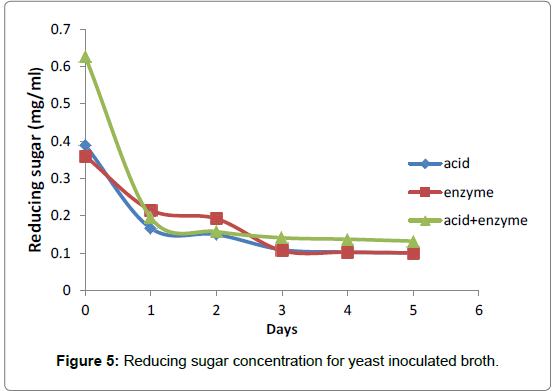
The algal biomass had initial sugar content of 0.39, 0.36 and 0.63 in mg/ml after acid, enzyme and combined acid and enzyme hydrolysis respectively. After 5 days fermentation at 30°C and pH 4.5 with initial yeast inoculum of 10%, the sugars were consumed rapidly resulting to increase ethanol concentrations while the sugar contents reduced to 0.10, 0.09 and 0.13 in mg/ml for acid, enzyme and co-hydrolyzed fermentation broths, respectively (Figure 5).
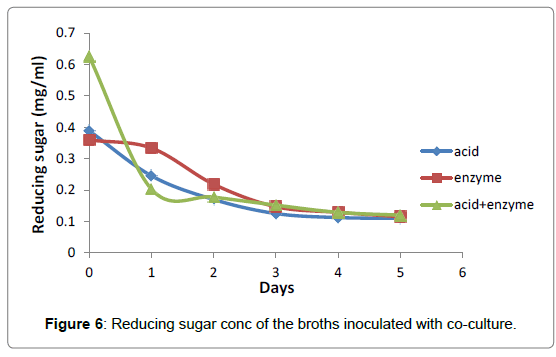
The fermentation broths inoculated with co-culture of A. niger and S. cerevisiae, the reducing sugars were consumed highly at co-hydrolyzed broth to 0.09, followed by acid then enzyme hydrolyzed fermentation broths reduced to 0.11 and 0.12 in mg/ml, respectively (Figure 6).
Ethanol yields of the fermentation
Ethanol concentration yields of the three saccharified fermentation broths resulted to steady increase from the beginning to the final day of fermentation. In acid and enzyme fermentation broths, ethanol concentration yields were lower than combined pretreated hydrolysate.
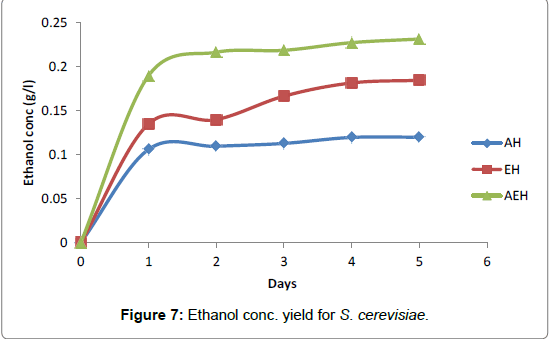
The fermentation broths using S. cerevisiae, the maximum ethanol yield recorded with combined acid and enzyme hydrolysate fermentation broth (0.23 g/l), directly followed by enzyme hydrolysate (0.18 g/l), while the least ethanol yield was obtained from acid hyrolysate (0.12 g/l) for the fermentation of 5 days at 30°C and pH 4.5 (Figure 7).
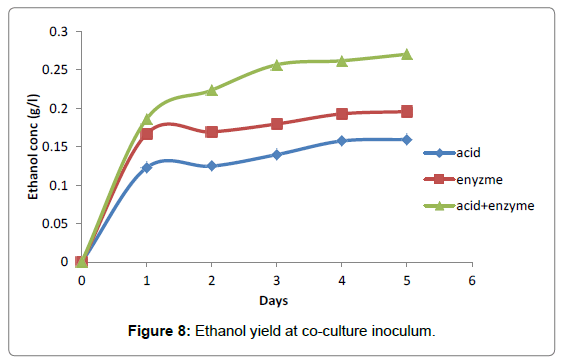
Co-culture of A. niger and S. cerevisiae used for the fermentation period resulted in the highest ethanol concentration yield (0.33 g/l) enzyme hydrolysate (0.19 g/l) while the least was obtained with acid hydrolysate (0.16 g/l) at 30°C and pH 4.5 (Figure 8).
Ethanol percentage in the distillates
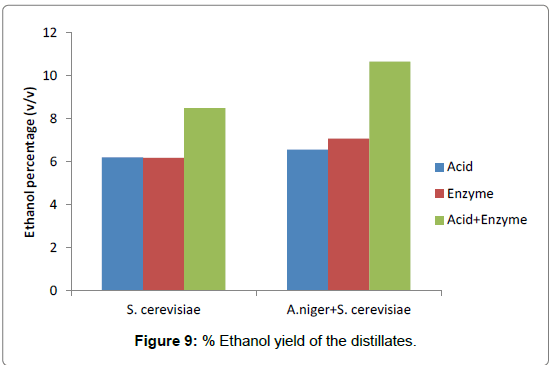
The study revealed that the maximum percentage ethanol yield for fermentation broths with the distillate of combined acid and enzyme hydrolysate inoculated with only S. cerevisiae obtained (8.50% v/v), followed by acid hydrolysate (6.21% v/v) while the least ethanol percentage yield from enzyme hydrolysate (6.18% v/v) (Figure 9).
Highest ethanol percentage of 10.66% v/v was obtained with combined acid and enzyme hydrolyzed fermentation broth, followed by enzyme hydrolysate (7.08% v/v) and acid hydrolysate as the least (6.56% v/v).
Discussion
In this study, six bacterial isolates were outstanding due to their ability to hydrolyze cellulose when used as a sole carbon source in the medium. Three different fungal species were selected due to their ability to elaborate amylolytic activity on the media. The isolate that showed the widest zone of clearance was selected as the highest amylolytic enzyme secreting fungi. The megablast search of 16S rDNA sequence of the selected isolates used for this study was identified to species level. The result obtained from the bacterial isolate (B30) showed an exact match for closely related sequences from the NCBI non-redundant nucleotide (nr/nt) database. The 16S rDNA of the isolate showed percentage similarity to other species at 99%. The Jukes-Cantor technique applied for computing distances among the evolutionary species were in agreement with the phylogenetic placement of the isolates 16S rDNA within the Bacillus sp. and revealed a close relatedness to Bacillus cereus R-14 (gi: 925175099) than other Bacillus sp. (Figure 3). The GenBank accession number of this isolate is Bacillus cereus-907-R KY198372. The PCR assays of fungal isolates were carried out using ITS1F and ITS4. The 18S rRNA sequence obtained from the fungal isolate (F16) produced an exact match during the megablast hunt for greatly related sequences from the NCBI non-redundant nucleotide (nr/nt) database. The 18S rRNA of the isolate showed percentage similarity to other species at 99%. The evolutionary gaps calculated using the Jukes- Cantor method were in agreement with the phylogenetic position of the 18S rRNA of the isolates within the Aspergillus sp. and revealed a close relatedness to Aspergillus niger isolate NJDL-12 (gi: 969987517) than other Aspergillus sp. (Figure 6). The Gen Bank accession number of this isolate is Aspergillus niger ITS-1 KY198375. The 18S rRNA sequence obtained from the fungal isolate (F14) produced an exact match during the megablast search for close related sequences from the NCBI non-redundant nucleotide (nr/nt) database. The 18S rRNA of the isolate illustrates percentage similarity to other species at 99%. The evolutionary gaps figured using the Jukes-Cantor method were in agreement with the phylogenetic position of the 18S rRNA of the isolates within the Saccharomyces sp. and revealed a close relatedness to Saccharomyces cerevisiae YJM681 (gi: 768856467) than other Saccharomyces sp. (Figure 3). The GenBank accession number of this isolate is Saccharomyces cerevisiae _ITS-1 KY198374.
Results on amylase enzyme production showed high amylase activity produced by solid state fermentation (SSF) by Aspergillus niger isolate NJDL-12 with plantain peel as the substrate. This is analogous to the study by Chimata et al. [31], where utmost extracellular amylase enzyme production (164 u/g) was recorded by SSF of wheat bran by a laboratory isolate Aspergillus sp. MK07. The main bottleneck in production of ethanol using biomass is the conversion process of starch and carbohydrate to fermentable sugars. Bioethanol production of ethanol is economically viable if the hydrolysis strategy such as enzyme production employed is optimized to reduce production cost which significantly lowers the bioethanol production cost to achieve current demand for biofuel. The results of this study demonstrated that cellulolytic and amylolytic enzymes cocktail preparation of Bacillus cereus strain R-14 and Aspergillus niger isolate NJDL-12, respectively can be used for microalgae biomass hydrolysis with greater advantages. The yield of the reducing sugars from its hydrolysis is lower than that from dilute acid hydrolysis indicating that saccharification activities of the enzyme are not adequate. It is likely that the activities of the amylase and cellulase enzymes were not sufficient for entire saccharification of microalgal cells [30,31]. Acid hydrolysis produced higher reducing sugar yields but lower ethanol yield than enzyme hydrolysis. However, reports showed that this may be as a result of some degradation by- products like furfural, hydroxyfurfural and other organic products produced by the acid pretreatment which may inhibit fermentation process [32]. Lower bioethanol yield at acid hydrolysate can also be as a result of higher salt concentration formed during pH adjustment which may inhibit growth and metabolism of the yeast [22]. But the enzymatic hydrolysates did not contain any inhibitory compounds as evidenced by the growth of the two organisms used for fermentation. This is similar to the study of production of cellulase from biomass feedstock and the enzyme used in saccharification of lignocelluloses for production of bioethanol [33]. The results show that integration of dilute acid and crude enzymes in the hydrolysis of microalgal biomass exhibited the best saccharification efficiency and highest yield of ethanol. This is as a result that the combined process has the capacity to decompose undisrupted cell wall carbohydrates of microalgal biomass. In addition salts formed after acid neutralization was harnessed by the crude enzyme cocktail in combined saccharification process prior to fermentation.
The microalgal biomass hydrolysates in this work were fermented by co-culture of Aspergillus niger isolate NJDL-12 and Saccharomyces cerevisiae YJM681 and S. cerevisiae alone. Co-culture fermentation by A. niger and S. cerevisiae yielded highest ethanol during the fermentation period than with their SHF process and highest percentage ethanol in the distillates was obtained using SHCF process. This may be due to various mixtures of sugars such as hexoses and pentoses released into the hydrolysates which cannot be utilized by S. cerevisiae but were hydrolysed to fermentable sugars by A. niger. This organism is capable of producing carbohydrate hydrolases and certain enzymes like amylases, cellobiases, xylanases which degrade the non-starch polysaccharides resulting to related increase in the amount of soluble sugars available in the fermentation broth [34-37]. The susceptibility of some sugars obtained after hydrolysis by A. niger during the fermentation process to the fermentable activity of S. cerevisiae became higher causing corresponding increase in ethanol yield. The result of this study agrees with the study production of ethanol by simultaneous saccharification and co-culture of A. niger and S. cerevisiae fermentation of yam peel which stated that most substrates were utilized for ethanol production in co-culture fermentation.
Conclusion
The different saccharification methods influence the concentration of reducing sugars available for bioethanol fermentation. Crude amylase and cellulase cocktail from local isolates of Aspergillus niger isolate NJDL-12 and Bacillus cereus strain R-14 when combined with acid hydrolysis were found to successfully produced high level of reducing sugars and bioethanol yield from microalgae biomass. Thus SHCF effectively produced higher ethanol yield than SHF.
Conclusively, the carbohydrate enriched microalgal biomass can be effectively hydrolyzed by integration of crude of enzyme cocktail of cellulase and amylase with dilute acid. Bioethanol production can be obtained from their hydrolysate. Thus, this provides a possible means to lessen the cost of biofuel production from microalgae.
References
- Jimoh SO, Ado SA, Ameh JB (2009) Simultaneous saccharification and fermentation of yam peel to ethanol by co-culture of Aspergillus niger and Saccharomyces cerevisiae. Biol J Trop 6: 99-100.
- Harun R, Jason WS, Cherrington T, Danquah M (2010) Microalgal biomass as cellulosic fermentation feedstock for bioethanol production. Renewable and Sustainable Energy Reviews.
- Zhu LD, Hiltunen E, Antila E, Zhong JJ, Yuang ZH, Wang ZM (2014) Microalgal biofuel: Flexible bioenergies for sustainable development. Renewable Sustainable Energy Reviews. 30: 1035.
- Markou G, Chatzipalidis I, Georgakakis D (2012) Carbohydrates production and bioflocculation characteristics in cultures of Arthrospira (spirulina) platenesis. Improvement through phosphorus limitation process. Bioenergy Resour 5: 915-925.
- Taherzadeh MJ, Karimi K (2007) Enzyme-based hydrolysis processes for ethanol from lignocellulosic materials: A review. Bio Resources 2: 707-738.
- Nagendran S, Hallen-Adams HE, Paper JM, Aslam M and Walton JD (2009) Reduced genomic potential for secreted plant cell wall degrading enzymes in the ectomycorrhizal fungus Amanita bisporigera, based on the secretome of Trichoderma reesei. Fungal Gen Biol 46: 427-435.
- Kumar A, Duhan JS (2011) Production and characterization of Amylase enzyme isolated from Aspergillus niger MTCC-104. Employing solid state fermentation. Inter J Pharm Biosci 2: 250-258.
- Pandey A, Nigam P, Selvakumar P, Soccol CR (1999) Solid state fermentation for the production of industrial enzymes. Curr Sci 77: 149-162.
- Kirankumar V, RaviSankar N, Shailaja R, Saritha K, Siddhartha E, et al. (2011) Purification and characterization of a-Amylase produced by Aspergillus niger using banana peels. J Cell Tissue Res 11: 2775-2780.
- Epegu CD (2006) Isolation of Microalgae from fresh water ponds at the African Regional Aquaculture Centre Aluu and the laboratory assessment of their economic potentials. Nig J Microbiol 20: 817-823.
- Agwa OK, Abu GO (2014) Utilization of poultry waste for the cultivation of Chlorella sp. for biomass and lipid production. Int J Curr Microbiol Appl Sci 3: 1036-1047.
- Teather RM, Wood P (1982) Use of Congo red polysaccharide interaction in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol 43: 777-780.
- Wilfinger WW, Mackey K, Chomczynski P (1997) Effect of pH and ionic strength on the spectrophotometric assessment of nucleic acid purity. Biotechniques 22: 478-481.
- Santou N, Nej N (1987) The neighbor-joining method: A new method for reconstructing phylogentic trees. Mol Biol Evol 4: 406-425.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725-2729.
- Miller GL (1956) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem, p: 426.
- Singh A, Olsen SI, Nigam PS (2011) A viable technology to generate third generation biofuel. J Chem Technol Biotechnol 86: 349-353.
- Kheng PP, Omar CI (2005) Xylanase production by local fungal isolates Aspergillus niger USM A11 via solid state fermentation using palm kernel cake as substrate. J Sci 27: 325-336.
- Miller T (1959) Use of DNS reagent for determination of reducing sugars. Anal Chem 31: 426-428.
- Miranda JR, Passarinho PC and Gouveia L (2012) Pretreatment optimization of Scenedesmus obliquus microalga for bioethanol production. Bioresour Technol 104: 342-348.
- Dhull N, Gupta K and Kumar S (2014) Heterotrophic and mixotrophic cultivation of Chlorella pyrenoidosa and the enzymatic hydrolysis of its biomass for the synthesis of third generation bioethanol. Peer J Preprints 2: e483v1.
- Salman JM, Ali MAM (2014) Bioethanol production from green alga Chlorella vulgaris under different concentrations of nitrogen. Asian J National Appl Sci 3: 27-36.
- Markou G, Angelidaki I, Nerantzis E and Georgakakis D (2013) Bioethanol production by carbohydrate-enriched of Arthrospira (spirulina) platensis. Energies 6: 3937-3950.
- Itelima J, Ogbonna A, Pandukur S, Egbere J, Salami A (2013) Simultaneous saccharification and fermentation of corn-cobs to bioethanol by co-culture of Aspergillus niger and Saccharomyces cerevisiae. Int J Environ Sci Dev 4: 329-342.
- Lee OK, Oh YK, Lee EY (2015) Bioehanol production from carbohydrate-enriched residual biomass obtained from lipid extraction of Chlorella sp. KR-1. Bioresour Technol 196: 22-27.
- Caputi A, Ueda M, Brown T (1968) Spectrophotometric determination of ethanol in wine. Am J Enol Vitic 19: 160-165.
- Oyeleke SB and Jibrin NM (2009) Production of bioethanol from guinea cornhusk and millet husk. Afr J Microbiol Res 3: 147-152.
- Riungu A, Salim AM, Njenga J, Yusuf AO and Waudo W (2014) Bioethanol production from waste crops and crop residues. J Appl Chem 7: 42-47.
- Gerken HG, Donohoe B and Knoshaug EP (2013) Enzymatic cell wall degradation of Chlorella vulgaris and other microalgae for biofuel production. Planta 273: 239-253.
- Kim KH, Choi IS, Kim HM, Wi SG and Bae HJ (2014) Bioethanol production from the nutrient stress induced microalgae Chlorella vulgaris by enzymatic hydrolysis and immobilized yeast fermentation. Bioresour Technol 153: 47-54.
- Palmarola-Adrados B, Choteborske P, Galbe M (2005) Ethanol production from non-starch carbohydrates of wheat bran. Bioresour Technol 96: 843-850.
- Sukumaran RK, Singhania R, Mathew GM, Pandey A (2009) Cellulose production using biomass feedstock and its application in lignocelluloses saccharification for bioethanol production. Renewable Energy 34: 421-424.
- Botella C, Diaz A, DeOry WC, Blandino A (2007) Xylanase and pectinase production by Aspergillus awamori on grape pomace in solid state fermentation. Process Biotechnol 42: 98-101.
- Akinyele BJ, Agbro O (2007) Increasing the nutritional value of plantain wastes by the activities of fungi using the solid state fermentation technique. Res J Microbiol 2:117-124.
- Mamma D, Kourtoglou E, Christak O and Poulos P (2008) Fungal multi enzyme production of industrial by-products of the citrus processing industry. Bioresour Technol 99: 2373-2383.
- Chimata MA, Sasidhar P and Challa S (2010) Production of extracellular amylase from agricultural residues by a newly isolated Aspergillus species in solid state fermentation. Afr J Biotecnol 153: 47-54.
- Jukes TH and Cantor CR (1969) Evolution of protein molecules. In: Mammalian Protein Metabolism. Academic Press, New York, pp: 21-132.
Citation: Agwa OK, Nwosu IG, Abu GO (2018) Saccharification and Bioethanol Fermentation of Carbohydrate-Extracted Microalgal Biomass by Genetically Identified Organisms. J Biotechnol Biomater 8: 279. DOI: 10.4172/2155-952X.1000279
Copyright: © 2018 Agwa OK, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 6945
- [From(publication date): 0-2018 - Dec 18, 2024]
- Breakdown by view type
- HTML page views: 6067
- PDF downloads: 878