Rice Root Genetic Research
Received: 01-Jul-2022 / Manuscript No. rroa-22-71010 / Editor assigned: 04-Jul-2022 / PreQC No. rroa-22-71010 (PQ) / Reviewed: 18-Jul-2022 / QC No. rroa-22-71010 / Revised: 22-Jul-2022 / Manuscript No. rroa-22-71010(R) / Accepted Date: 22-Jul-2022 / Published Date: 29-Jul-2022 DOI: 10.4172/2375-4338.1000312
Abstract
Rice is one of the most important cereal crops, feeding more than 50% population of the world. To meet the demand of increasing population, rice production has to be improved continually. As a very important part of rice plant, root system plays multiple roles in rice growth: anchorage of the plant, acquisition of water and nutrient elements, and biosynthesis of amino acids and hormones, etc. Almost all of the hot spots about rice research are associated with rice root: drought tolerance, lodging resistance, and efficient use of nutrition, the goal is to increase the grain yield with desirable seed quality. Although the understanding about rice root has been expanded in the last decades, there remain much to be done about root morphology and physiology, especially in root genetics. Rice root research is an exciting and focusing field in recent years.. It has several characteristics, such as high cellulose and hemicelluloses content that can be readily hydrolyzed into fermentable sugars. But there occur several challenges and limitations in the process of converting rice straw to ethanol.
Keywords: Rice (Oryza sativa L.); Root; Genetic research;
Keywords
Rice (Oryza sativa L.); Root; Genetic research
Introduction
Rice is one in all the foremost vital food crop, feeding over half the world’s population. The increasing population and economic development are move a growing pressure for increase in food production. Meanwhile, rice yield increase eventually slowed in China likewise as in different countries. The typical yearly increase in yield born from three.7% in Eighties to zero.9% in Nineties to full fill the demand of food for increasing population, raising the yield ceiling of rice remains a priority task for rice breeders. Roots sense and response to abiotic and communicate with the shoot via communication pathway. Hormones play a important role in root/shoot communications. ABA (abscisic acid), ethene and growth regulator function the signals for communication between root and shoot in rice. Roots will regulate not solely stomatal electrical phenomenon, and conjointly have an effect on the posture of leaf and chemical change rate below soil resistivity, nutrient, drought and salt stresses. Importance of root genetic improvement It is the roots that absorb most nutrients and water (Russell, 1977). Among essential nutrient parts needed for rice growth, inorganic carbon is absorbed principally by leaves within the variety of dioxide, the opposite essential mineral parts square measure all absorbed principally through root surface from the soil. Root is that the foundation of rice development. As delineate within the report by he high grain yield was principally because of a bigger sink size (total range of spikelets) as a results of a bigger raceme. The low proportion of stuffed grains was closely related to a fast shrivelled root activity throughout grain filling. Additional analysis is required to know the mechanism concerned within the low proportion of stuffed grains and yield fluctuation and to boost the yield performance in elite hybrid lines. It is the roots that absorb most nutrients and water essential nutrient parts needed for rice growth, inorganic carbon is absorbed principally by leaves within the variety of dioxide, the opposite essential mineral parts square measure all absorbed principally through root surface from the soil. Root is that the foundation of rice development. As delineate within the report by Zhang et al. (2009), the high grain yield was principally because of a bigger sink size (total range of spikelets) as a results of a bigger raceme. The low proportion of stuffed grains was closely related to a fast shrivelled root activity throughout grain filling. additional analysis is required to know the mechanism concerned within the low proportion of stuffed grains and yield fluctuation and to boost the yield performance in elite hybrid lines.
Materials and Method
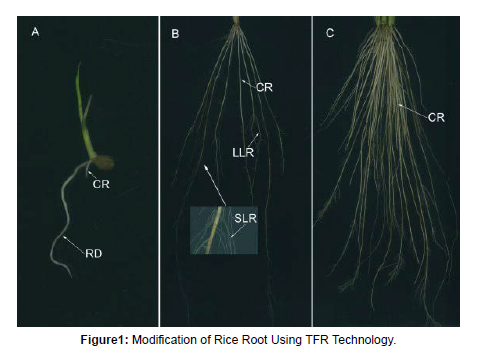
Drought is one in all the foremost severe abiotic stresses limiting rice productivity within the world, and poses a significant threat to the property of rice yields in rain fed agriculture. Development of drought resistant rice is one in all the objectives within the watersaving agriculture programs. Acquisition of a lot of water from soil may be a mechanism for drought tolerance in rice. Nitrogen use potency (NUE) for cereal production is more or less thirty third worldwide. The remaining N from plant food is lost to the atmosphere or leached into the groundwater and different fresh bodies , that is inflicting serious N pollution and turning into a threat to international ecosystems N pollution is currently claimed the third major threat to our planet when multifariousness loss and climate change Rice bears a shallow scheme that’s comprised of 1 seminal root (radicle), varied accidental roots (crown roots) arising from consecutive nodes, and enormous and little lateral root rising from primary roots (Figure 1) Dissecting genetic and molecular mechanisms dominant rice root development is essential for the event of recent rice hat square measure higher custom-made to adverse conditions and for the assembly of sustainably achieved rice yield potential Early in Eighties, and have created.
Data Analysis
There are four reported genes related to radicle development in rice. The ral1 is the first mutant impaired in both procambium development and vascular patterning to be isolated in a mono species, which produce normal Crown rootless1 (crl1) mutant is defective in crown root formation (Inukai et al., 2005). It showed auxin-related abnormal phenotypic traits in the roots, such as decreased lateral root number, auxin in sensitivity in lateral roots (LRs) formation, and impaired root gravitropism/ whereas aboveground organs were normal. ARL1 is the same gene with CRL1 (CRL1 encodes a protein with a LOB domain. It expresses in lateral and adventitious root primordial tiller primordia, vascular tissues scutellum, and young pedicels. CRL2 is involved in crown root formation, the initiation and subsequent growth of adventitious roots primordia of mutant are impaired (Inukai et al., 2001). CRL3 is exclusively involved in the formation of the crown root primordia, but not in the formation of other types of root primordial and shoot apical meristem. CRL4 encodes OsGNOM1, which expressed in AR(adventitious root) primordia, vascular tissues, LRs, root tips, leaves, anthers and lemma veins . CRL4 is mediated by OsPINs family, such as OsPIN2, OsPIN5b, OsPIN9, and affected the formation of ARs through regulating PAT (polar auxin transport) (CRL5 encodes a member of the large AP2/ERF transcription factor family protein. The auxin-induced CRL5 promotes crown root initiation through repression of cytokinin signaling by positively regulating type-A RR, OsRR1.Root elongation is essentially driven by stem cells localized in apical meristems of roots. The mutants rrl1, rrl2 both show shorter root, RRL1, RRL2 inhibit the maintenance of root apical meristem and cell elongation The mutant rrl3 with short roots is highly sensitive to mechanical stimulus. RRL3 specifically regulates the cell production process in the root meristematic zone under mechanically impeded condition, and does not regulate the sensitivity to ABA (abscisic acid), IAA (indoleacetic acid) and ethylene. SRT1 is a short root gene.
Result and Discussion
There are three transport systems for the uptake of in plants roots: two high affinity transport systems (HATS: constitutive HATS, cHATS, and inducible HATS, iHATS) and one low-affinity transporter system (LATS), and two for uptake of , one HATS and one LATS. Ammonium in comparison to nitrate as N source for rice promoted uptake and translocation of total N from root to shoot and resulted in higher shoot yield. Supply of the two forms of N showed the highest biomass yield of both roots and shoots and total N The uptake and distribution of P in plants requires multiple P transport systems throughout growth and development. considered that an effective adaptation for enhancing P uptake was to increase root growth mapped a major QTL, pup1, for P uptake to a 13-cM marker interval on the long arm of chromosome 12. characterized this gene and named PSTOL1, it is the first phosphorus-efficiency gene identified in plants. The gene encodes enzyme that significantly enhances grain yield from rice plants grown on phosphorus-deficient soils.The PHT1 and PHT2 families in plant function absorption and transportation of P. Thirteen putative high-affinity P transporter genes belonging to the Pht1 family (OsPT1– OsPT13) have been identified in the rice genome (OsPT1is involved in the OsPHO2-regulated P pathway (OsPT1 is a key member of the Pht1 family involved in P uptake and translocation under P-replete conditions. Expression of OsPT13 is constitutively independent of P supply. OsPT2 is a low-affinity P transporter, expressed abundantly in P-starved roots and facilitate the transport of P from roots to shoot. OsPT6 plays a broad role in P uptake and translocation throughout the plant). OsPT9 and OsPT10 express in the root epidermis, root hairs and lateral roots, with their expression being specifically OsPT11 and OsPT13, are exclusively induced in roots by inoculation with arbuscular mycorrhiza fungi. OsPT11 activation is independent of the nutritional status of the plant and phosphate availability in the rhizosphere, OsPHT2;1, a putative low affinity phosphate transporter gene, is involved in the P accumulation in leaves and P translocation through plants as showed in (Table 1).
| Phenotype | Gene | Locus ID |
|---|---|---|
| Fe utilization | OsbHLH133 | LOC_Os12g32400 |
| OsFRDL1 | LOC_Os03g11734 | |
| OsIRO2 | LOC_Os01g72370 | |
| OsIRT1 | LOC_Os03g46470 | |
| OsNAS1 OsNAS2 OsNAS3 |
LOC_Os03g19427 LOC_Os03g19420 LOC_Os07g48980 |
|
| NAAT1 | LOC_Os02g20360 | |
| OsYSL2 OsYSL15 OsYSL16 OsYSL18 |
LOC_Os02g43370 LOC_Os02g43410 LOC_Os04g45900 LOC_Os01g61390 |
|
| Si acquisition | LSI1 | LOC_Os02g51110 |
| Cd accumulation | OsHMA2 OsHMA3(qCdT7) OsHMA9 |
LOC_Os06g48720 LOC_Os07g12900 LOC_Os06g45500 |
| Water acquisition | OsPIP1,2,4,5,7; OsTIP4;1 |
|
| Abiotic stress tolerance | MAIF1 | |
| OsMSR2 | LOC_Os01g72530 |
Researches on aquaporins in rice mostly focused on PIPs. Thirtythree genes for aquaporins in rice have been identified, of which six genes, including OsPIP2;4 and OsPIP2;5, express predominantly in roots. 14 genes, including OsPIP2;7 and OsTIP1;2, are found in leaf blades. Eight genes, such as OsPIP1;1 and OsTIP4;1, express in leaf blades, roots and anthers (Sakura et al., 2005). There are 10 rice PIP genes (OsPIPs) that are classified into two subgroups (OsPIP1 and OsPIP2), of which three members OsPIP1–3, OsPIP2–2, and OsPIP2–7 are root specific in seedlings (Guo et al., 2006). Since Hales carried out root research with excavation method as early as 1727, a number of approaches, such as root box, pin-board, minirhizotrons, CT scans, gel-based imaging have been improved for root studies. Recent years, computer assistant image analysis have been developed quickly (see Root Methods, Smit et al., 2000). The synchrotron X-ray computed tomography has been used as a noninvasive method to observe how aerenchyma develops from rice primary root (Karahara et al., 2012). A three-dimensional imaging technique allows to perform a quantitative morphological analysis and time-course, and also in-situ observations of aerenchyma formation to phenotype root traits during seedlings.
Conclusion
However, many methods are still time-consuming and laborious, and largely influenced by the complex underground environments.
Root sampling procedures are often destructive. It is impossible to sample intact root system from plants in field environments. Soilless culture techniques have provided a simple and convenient methods with which the whole root system could be extracted from the plants. But the soilless culture system could not completely mimic the environments of paddy field, thus the information obtained in soilless culture usually do not exactly reflect root feature under natural conditions. For identifying and screening the root traits, more facilitated and effective methods, especially the large-scale screening techniques for root measurement in paddy field are urgently required. Quantitative trait loci (QTL) mapping is a major approach for investigating complex genetic traits such as root, but QTL were not fine-mapped with appropriate selectable markers, the desired gene might have been lost in the selection process (Gowda et al., 2011). To cope with this problem, association mapping(meta-analysis) as a promising method was introduced to genetic dissection of complex traits. Using association mapping, it is possible to locate QTLs with better precision than using a mapping population (Courtois et al., 2009, Courtois et al., 2013). Among the large number of root QTLs identified in past decades, few major QTLs have been cloned and introgressed into another background (Shen et al., 2001). Introduction of DRO1 into a shallow-rooting rice cultivar was a successful practice. It enabled the resulting line to avoid drought by increasing deep rooting, which maintained high yield performance under drought conditions relative to the recipient cultivar(Uga et al., 2013)UBMERGENCE 1 (SUB1) is a robust quantitative trait locus from the submergence tolerant FR13A landrace. The marker-assisted introgression of the SUB1 region has successfully improved submergence tolerance in a wide range of megavarieties without any penalties on development, yield, and grain quality (Bailey-Serres et al., 2010). Compared with QTL mapping, exploring beneficial genes with root mutants is a more efficient approach, and a number of genes reported are explored through root mutants. In addition, a number of genome-wide large-scale studies have been performed (Takehisa et al., 2012, Zhai et al., 2013). These provide researches useful techniques to unveil molecular mechanisms of root development. For example, transcriptome analysis of rice mature root tissue and root tips at two time points identified 1761 root-enriched transcripts and 306 tip-enriched transcripts involved in different physiological processes.
References
- Roslin NA, CheYa NN, Sulaiman N, Alahyadi LAN, Ismail MR, et al. (2016) Mobile Application Development for Spectral Signature of Weed Species in Rice Farming. Pertanika J Sci Technol 29: 2241-2259.
- Dilipkumar M, Chuah TS, Goh SS, Sahid I (2020) Weed management issues, challenges, and opportunities in Malaysia. Crop Prot 134: 104-123.
- Giacomo R, David G (2017) Unmanned Aerial Systems (UAS) in Agriculture: Regulations and Good Practices. In E-Agriculture in Action: Drones for Agriculture; FAO: Roma, Italy.
- Roslin NA, CheYa NN (2020) M.R. Smartphone Application Development for Rice Field Management Through Aerial Imagery and Normalised Difference Vegetation Index (NDVI) Analysis. Pertanika J Sci Technol 29: 123-129.
- Sulaiman N, CheYa NN, Mohd Roslim MH, Juraimi AS, Mohd NN, et al. (2020) The Application of Hyperspectral Remote Sensing Imagery (HRSI) for Weed Detection Analysis in Rice Fields: A Review Appl Sci 12: 2570-2580.
- Wang S, Gao (2018) Improved 93-11 genome and time-course transcriptome expand resources for rice genomics. Front Plant Sci 12: 769-780.
- Wang W(2018) Genomic variationin 3010 diverse accessions of Asian cultivated rice. Nature 55: 43–49.
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, et al. (2007) The TIGR Rice Genome Annotation Resource: Improvements and new features. Nucleic Acids Res 35: D883–D887.
- Sakai H, Lee SS, Tanaka T (2004) RiceAnnotation Project DataBase (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol 54: e64-e65.
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Citation: Ibro G (2022) Rice Root Genetic Research. J Rice Res 10: 312. DOI: 10.4172/2375-4338.1000312
Copyright: © 2022 Ibro G. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 1742
- [From(publication date): 0-2022 - Mar 12, 2025]
- Breakdown by view type
- HTML page views: 1466
- PDF downloads: 276