Research Article Open Access
Ribosomal Protein S1: An Important Trans-Translational Factor
| Iwona K. Wower1, Nusrat Jahan2, Christian Zwieb3 and Jacek Wower1* | |
| 1Department of Animal Sciences, 210 Upchurch Hall, Auburn University, Auburn, AL 36849-5415, USA | |
| 2Department of Molecular Genetics and Microbiology, Life Sciences Building, Stony Brook University, Stony Brook, NY 11794-5222, USA | |
| 3University of Texas Health Science Center at San Antonio, 7703 Floyd Curl Drive, San Antonio, Texas 78229, USA | |
| *Corresponding Author : | Jacek Wower Department of Animal Sciences 210 Upchurch Hall, Auburn University Auburn, AL 36849-5415, USA Tel: +1 334 844 1508 Fax: +1 334 844 1519 E-mail: wowerja@auburn.edu |
| Received December 01, 2012; Accepted December 27, 2012; Published December 31, 2012 | |
| Citation: Wower IK, Jahan N, Zwieb C, Wower J (2013) Ribosomal Protein S1: An Important Trans-Translational Factor. Biochem Physiol S2:001. doi:10.4172/2168-9652.S2-001 | |
| Copyright: © 2013 Wower IK, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Biochemistry & Physiology: Open Access
Abstract
S1 is an essential protein in Escherichia coli. Although not present in all bacteria, its importance for the initiation and elongation stages of protein synthesis is well established. Beside its roles as a ribosomal protein, S1 promotes transcriptional cycling, regulates bacteriophage T4 gene expression, forms a complex with the phage. λ protein β involved in recombination, and is a subunit of the fr and Qβ RNA bacteriophage replicases. Protein S1 was also shown to bind to tmRNA, an essential component of trans-translation. Although the physiological significance of protein S1 for trans-translation has been debated for many years, recent studies clearly demonstrate that protein S1 constitutes an important, yet poorly understood component of trans-translation. We show that binding of protein S1 to the free tmRNA is a prerequisite for the association between tmRNA and stalled ribosome. S1 transits the ribosome together with the tmRNA as defective proteins are targeted for proteolysis. These findings establish protein S1 as an important target for pharmacological intervention.
| Keywords |
| Ribosomal protein S1; Trans-translation; tmRNA; SmpB |
| Introduction |
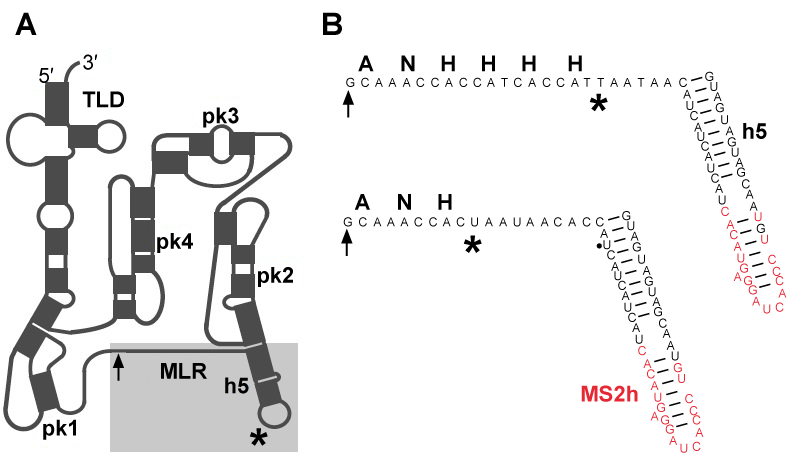
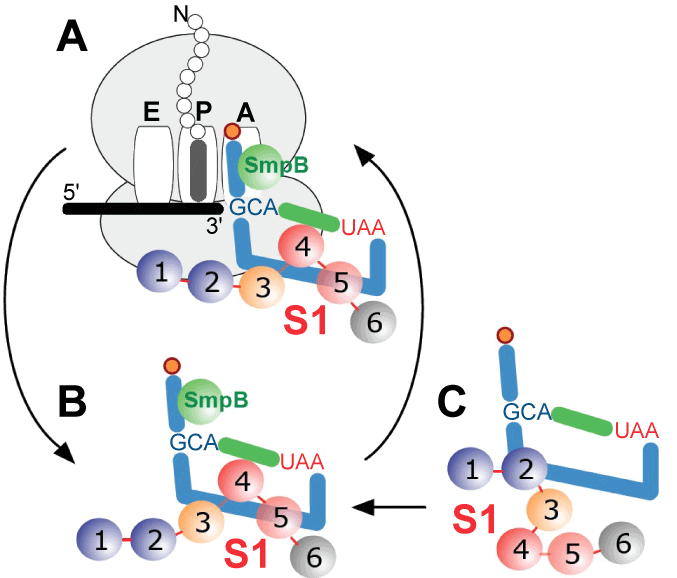
| When the ribosome stalls on a mRNA molecule that lacks a stop codon, bacteria use transfer-messenger RNA (also known as 10S RNA or SsrA) and a Small protein B (SmpB) to recycle the ribosome, tag the defective protein for proteolytic degradation and accelerate degradation of the truncated mRNA [1,2]. Comparative analyses of tmRNA sequences demonstrated that tmRNA is composed of tRNA-like domain and mRNA-like region, denoted in figure 1A as TLD and MLR respectively. In E. coli tmRNA, TLD and MLR are connected by four pseudoknots (pk-pk4) [3-5]. Three to six pseudoknots may be present in other bacterial tmRNAs. Within the TLD-SmpB complex, SmpB mimics the anticodon arm of a canonical tRNA [6]. The C-terminal tail of SmpB has been shown to mimic mRNA, and play an essential role in the functioning of ribosome-bound tmRNA [7,8]. |
| Most information about trans-translation comes from studies using E.coli as the model organism [9]. Trans-translation is initiated by binding of tmRNA:SmpB complex to the stalled ribosome. As the TLD:SmpB module moves from the A to the P, and then to E site of the ribosome, the MLR is translated into a peptide, which constitutes a relatively promiscuous signal that is recognized by a number of proteases. The tagging of the defective protein ends when the stop codon, which marks the 3’ end of MLR, enters the A site and triggers termination. TmRNA functions are enabled by canonical translational factors. After aminoacylation by alanyl-tRNA synthetase, tmRNA forms a complex with GTP and Elongation Factor Tu (EF-Tu) that delivers it to the ribosomal A site. Translocation of tmRNA from the A site to the P site, and then to the E site is mediated by Elongation Factor G (EF-G) [10]. |
| We discovered that ribosomal protein S1 binds to both free and ribosome-bound tmRNA [11]. As protein S1 is not found in all bacteria, the physiological significance of protein S1 binding to tmRNA was frequently debated [12-14]. However, recent studies clearly demonstrate that protein S1 facilitates trans-translation in a way that is distinctly different from its functions as a ribosomal protein during canonical translation. Noteworthy is that protein S1 binds to tmRNA before its association with the ribosome [15]. This interaction is inhibited by the tuberculosis drug pyrazinamide and prevents the tmRNA from binding to stalled ribosomes, and thus the tagging of immature protein [16]. In the present study, we demonstrated that S1 remains bound to tmRNA as it transits the ribosome, and that protein S1 is likely to interact with a large region of tmRNA, including the MLR and pseudoknots pk2 and pk3. At present, it is not clear whether protein S1 remains bound to tmRNA, after termination of trans-translation. It is also not clear what specific roles the domains of protein S1 play in trans-translation. To initiate discussion and stimulate future studies, we review here the structural and functional data that are pertinent to the understanding of interactions between protein S1 and tmRNA, both on and off the ribosome. |
| Materials and Methods |
| Preparation of mutant MS2-tmRNA-4 and MS2-tmRNA-7 |
| MS2-tmRNA(H8), a variant of E. coli tmRNAH8 that can bind the coat protein from MS2 bacteriophage, was constructed by PCR-directed mutagenesis, as described earlier [17]. Two mutants, called MS2-tmRNA-4 and MS2-tmRNA-7 were prepared (Figure 1B). |
| Protein tagging by MS2-tmRNA-4 and -7 |
| E. coli strain IW363 was transformed with plasmid pETrpmA-At-3, which carries one copy of an rpmA gene encoding a truncated ribosomal protein L27, one copy of SmpB gene and one copy of modified ssrA gene, encoding either MS2-tmRNA-4 or -7 [17]. Synthesis of truncated ribosomal protein L27 and following tagging activity was induced by addition of 1 mM IPTG to logarithmically growing cells. The liquid culture was incubated at 37°C for 3 hr. The expression of truncated protein L27 and its tagging were analyzed by SDS-Polyacrylamide Electrophoresis (SDS-PAGE) [17]. |
| Purification of MS2-tmRNA:Ribosome Complexes |
| IW363 cells carrying pETrpmA-At-3 were grown at 37°C to an A600 of 0.2 in 2xYT broth supplemented with kanamycin (50 μg/ml), chloramphenicol (30 μg/ml) and ampicillin (200 μg/mL), before adding 1 mM IPTG. When the cell culture reached an A600 of 0.8, cells were pelleted by centrifugation, resuspended in 100 mL of lysis buffer (20 mM Tris-HCl pH 7.5, 150 mM KCl, 2mM DTT and 5% glycerol) containing RNase-free DNase I (Boehringer Manheim) at 3 μg/ml, and lysed in a French press. Lysates were clarified by a 15 minutes long centrifugation at 14,000 g at 4°C. Supernatants were layered in portions of 16 mL on 9 mL of 1.1 M sucrose in 20 mM Tris-HCl pH 7.5, 100 mM NH4Cl, 10 mM Mg(CH3COO)2 and 3 mM β-mercaptoethanol (binding buffer) to isolate crude ribosomes. After a 16 hr-long centrifugation for at 33,000 rpm in a Beckman Ti 50.2 rotor, the ribosomes were dissolved in 2 mL of binding buffer. To isolate MS2-tmRNA-4 and -7:ribosome complexes, 1.3 mg GST-MS2 fusion protein was mixed with 20 mg of crude ribosomes and loaded onto a 1.5 mL Glutathione-Sepharose column (Amersham Biosciences). The column was washed with 10 mL binding buffer and ribosomes were eluted with 3 mL of elution buffer (20 mM Tris-HCl pH 7.5, 100 mM NH4Cl, 10 mM Mg(CH3COO)2 and 10 mM reduced glutathione). The eluted ribosomes were layered on 9 ml of 1.1 M sucrose in binding buffer, and centrifuged for 16 hr at 33,000 rpm in a Beckman Ti 50.2 rotor. This step was necessary to remove free MS2-tmRNA-4:SmpB complexes, which co-purify with MS2-tmRNA-4 and -7:ribosome complexes, during affinity chromatography step. The tmRNA:ribosome complexes were resuspended in 1 mL of binding buffer and stored at -80°C. Protein composition of the purified tmRNA:ribosome complexes was analyzed by SDS-PAGE, as described earlier [17]. |
| PCR site-directed mutagenesis |
| Site-directed mutants were constructed using PCR with a single mutagenic primer [18]. 20 mL step 1 PCR reactions were set up in a 250 μL polypropylene strip (Nunc). Each sample contained 2 mM of mutagenic primer and the upstream oligonucleotide a, a PCR reaction buffer (50 mM Tris-HCl pH 8.3, 0.5 mg/mL Bovine Serum Albumin (BSA), 1% (v/v) Ficoll, 1 mM Cresol Red 1), 3 mM MgCl2, 200 mM each dNTP, 10 ng of pT710S#21, and 2.5 units of Taq polymerase (GibcoBRL, Life Technologies). The reaction mixtures were transferred to 50 mL capacity borosilicate glass capillaries (Idaho Technology Inc.), and the capillaries were closed by melting the ends using a cigar lighter. The PCR was carried out in a Rapidcycler PCR machine (Idaho Technology Inc.), with 30 cycles of denaturation at 94°C, 15 sec annealing at 45°C, and 15 sec polymerization at 72°C. After the incubation, the ends of the capillary were broken, samples were transferred to the polypropylene strip, mixed with 4 mL loading buffer (0.25% (w/v) bromophenol blue, 0.025% (w/v) xylene cyanol FF, 30% glycerol (v/v) in 0.5 X TAE), and analyzed by electrophoresis on 2% (w/v) agarose gels. The gel was placed on a 365 nm long-wavelength ultraviolet transilluminator and the desired fragments were excised from the gel with a clean razor blade. A 0.5 mL microfuge tube with a small hole at the bottom was placed into a 1.5 mL collection tube. Whatman 3 MM filter paper-mache soaked in TE was applied to cover the hole, and extra liquid was removed by a quick spin. The gel was transferred into the 0.5 mL microfuge tube, and the DNA was collected by a 15 min centrifugation at 15,000 rpm. |
| Step 2/3 PCR reaction mixtures of 100 mL were assembled with 2 mL of extracted DNA from the first-step PCR, 50 mM KCl, 10 mM Tris⋅HCl pH 8.3, 250 mM of each dNTP, 10 ng of pT710S#21 DNA, 1.5 to 3 mM MgCl2, and 5 units of Taq polymerase. A single thermal-cycle extension of 5 min at 95°C, 2 in at 37°C and 10 min at 72°C was carried out (step 2) in a GeneAmp PCR System 2400 (Perkin-Elmer Cetus). 0.4 mM each of the upstream oligonucleotide b and the downstream oligonucleotide c were added, and PCR was carried out for 25 cycles (step 3) consisting of 30 sec at 95°C, 2 min at 60°C and 2 min at 72°C. 5 mL aliquots of the PCR product were analyzed by electrophoresis on 1.5% agarose gels. |
| Phenol/chloroform extracted and ethanol precipitated DNAs (see above) obtained in step 2/3 and the pT710S#21 vector DNA were digested with the restriction enzymes EcoRI and HindIII. The digested DNAs from the digestion reactions were extracted with phenol/chloroform and precipitated with ethanol (see above), and analyzed by electrophoresis on 1.5% agarose gels. The purified linearized vector pT710S#21 and mutant DNAs of expected length were extracted from the gel, using the 3 MM paper spin method described above. |
| To estimate the DNA concentrations of vector and insert for ligation diluted DNA samples were electrophoresed on 1.5% agarose gels. A 20 mL ligation reaction contained approximately 50-90 ng of insert DNA and 200 ng of purified linearized vector, in 1X T4 DNA ligase buffer (50 mM Tris-HCl, 10 mM MgCl2, 10 mM DTT, 1 mM ATP, 50 mg/mL BSA, pH 7.8), with 400 U of T4 DNA ligase (New England Biolabs). Samples were incubated overnight at 15°C. For the transformation of the ligated DNA, 10 mL aliquot of the ligation reaction were transferred to a 1.5 mL microfuge tube on ice, containing 100 mL of competent E. coli DH5α cells (Library Efficiency, Gibco BRL, Life Technologies), and mixed gently by tapping the tube with the fingers. After a 30-min incubation on ice, the mixture was heated at 42°C for 40 sec, and then returned to ice for 2 min. One millilitre of room temperature SOC medium (GibcoBRL, Life technologies) was added and the sample was incubated at 37°C for one hour, with shaking at 225 rpm. 100 mL of sample were plated on LB plates containing 200 mg/mL Ampicillin. The main portion of the cells was concentrated by centrifugation at 3,200 rpm for 10 min in a Sorvall RT 6000B, and the cells in the pellet were plated on Ampicillin containing LB plates, and incubated overnight at 37°C. |
| The names and sequences of the deoxyoligonucleotides used in the PCR reactions were as follows: |
| a. 5’-TCCTTAATCTTCCCCTCCGTAGGCACCCCAGGCTTTACACT- 3’; |
| b. 5’-TCCTGAATCTTCCCCTCCGT-3’; |
| c. 5’-CGCCAGGGTTTTCCCAGTCACGAC-3’; (m1) 5’-CTGCTTAGAGCxTAAGCATGTAG- 3’; (m2) 5’-ATCAGGCTAxTAAGCATGTAG- 3’; (m3) 5’-CTGCTTAGAGCxATCGCGTGGA- 3’; (m4) 5’-CCCAAAAGAGxGTTTGTTAGTG-3’; (m5) 5’-CCCAAAAGAGxTAAGCATGTAG-3’; (m6) 5’-CTGCTTAGAGCGTTTTTAGTG- 3’; (m7) 5’-AAGCCGCAAAAAxCCTCTCTCCCT- 3’; (M8) 5’-AAGCCGCAAAAAxATCGCGTGGA- 3’; (m9) 5’-AAGCCGCAAAAAxGTTTGTTAGTG-3’. |
| An “x” in the primer sequence indicates where nucleotides were deleted. |
| Formation of RNA:protein S1 complexes |
| Interactions between E. coli protein S1 and mutant tmRNAs were tested by gel mobility shift assay. A standard 20 mL reaction contained 0.25-1.0 mg of RNA, 0.1-2.0 mg of protein S1 in 10 mM Tris-acetate (pH 7.6), 100 mM NH4Cl, 10 mM Mg-acetate, 1 mM DTT, 0.02% NP-40, 0.1 mg BSA/mL, and 5% (v/v) glycerol (binding buffer). RNA samples with appropriate dilutions were prepared in RNase-free water and aliquoted in 1.5 mL microfuge tubes. After 10 min of incubation at room temperature, samples were placed on ice for 10 to 30 min, and then loaded onto a preelectrophoresed native 5% polyacrylamide gel. Electrophoresis was carried out 3 hours at 4°C. Gels were stained with 10 mg/mL ethidium bromide in 40 mM Tris-acetate and 2 mM EDTA (pH 8.0), and visualized under ultraviolet light. Gel pictures were taken and ImageJ software (available at http://rsb.info.nih.gov/ij/) was used to determine the pixels in each band. The command Analyze/Gels/Plot Lanes was used to obtain the profile plot of each lane. Base and drop lines were drawn to define closed areas. The number of pixels in each area was measured with the wand tool. The degree of binding was determined either by measuring the formation of complex (% of binding=(complex/total RNA)×100), or by measuring the disappearance of free RNA (% of binding=(free RNA/total RNA)×100). |
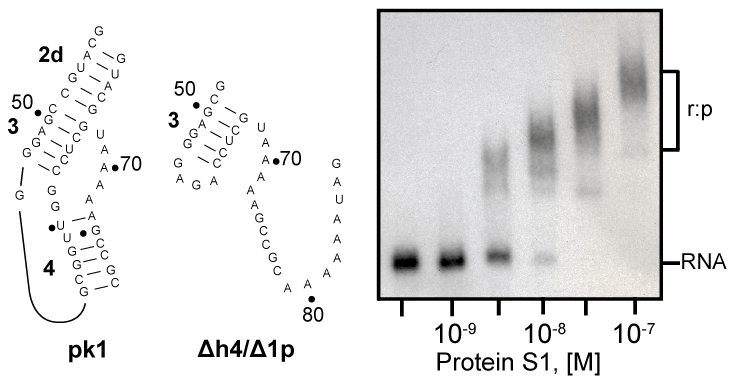
| Preparation of pk1 and Δh4/Δ1p RNAs |
| Both RNAs were synthesized by in vitro transcription in reactions containing 50 mM Tris-HCl (pH 8.1), 50 mM NaCl, 2.5 mM each NTP, 30 mM MgCl2, 1 mM spermidine, 5 mM DTT, 0.05 mg/mL T7 RNA polymerase and synthetic DNA templates [19]. RNA transcripts were labeled at their 3’ ends with [5’-32P]pCp and RNA ligase, and then purified on a denaturing 10% polyacrylamide gel in 100 mM Tris-100 mM H3BO4 (pH 8.3), containing 2.5 mM EDTA and 8 M urea [20]. |
| Results and Discussion |
| Structure of protein S1 in Gram-negative and Gram-positive bacteria |
| Protein S1 has been identified in a wide variety of bacterial groupings, including the Gram-positive and Gram-negative [21]. In E. coli, S1 is the largest of the ribosomal proteins (M.W.-61 kDa), consisting of 557 amino acid residues. It has six repeating homologous OB (oligonucleotide-oligosaccharide-binding) folds, or S1 domains, and usually denoted as R1-R6. The OB fold is found also in a number of other RNA-associated proteins. Insights into the structure of the S1 domains were provided using NMR, initially by Bycroft et al. [22], who determined the structure of the S1 domain of the E. coli polynucleotide phosphorylase, and more recently by Salah et al. [21], who reported the solution structure of the R4 and R6 domains of the E. coli protein S1. |
| A number of studies demonstrated that E. coli ribosomal protein S1 has two functional regions. The N-terminal region, composed of domains R1 and R2 (amino acid residues 1-193), is necessary for assembly of the Qβ replicase, and is also involved in the binding of protein S1 to ribosomes [23,24]. While other ribosomal proteins are strongly bound to ribosomal RNAs, the association of protein S1 with the ribosome is reversible, and depends on protein:protein interactions. Interestingly, using truncated versions of E. coli ribosomal protein S1, R2 was found to be essential for the recognition of tmRNA [25]. The second region of protein S1 consists of domains R4 through R6 that participate in the binding of mRNAs, and many other natural and synthetic RNA molecules. These interaction are not necessarily strictly sequence specific, as they can involve the U-rich sequences in the leader of bacterial mRNAs, as well as poly(rC) and poly(rA) [26]. |
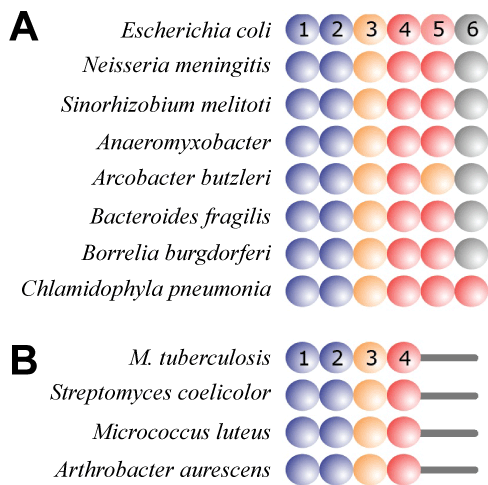
| Because protein S1 was believed to be present only in a selected group of Gram-positive bacteria, while tmRNA and SmpB had been identified in practically all bacteria, the involvement of protein S1 in trans-translation has been questioned in the past [12-14]. However, recent structural studies have identified S1 or S1-like proteins in nearly all bacterial groupings [21] (Figure 2). Because the binding of protein S1 to tmRNA is required for trans-translation in the Gram-positive Mycobacterium tuberculosis, one can expect that proteins S1 found in the related Actinobacteria (Figure 3) will be able to support trans-translation [16,21]. Interestingly, it has been shown Streptomyces coelicolor tmRNA plays an essential role in providing instantaneous response to environmental stressors [27]. An important case for the discussion on the involvement of protein S1 in trans-translation has been recently reported by Takada et al. [14]. According to this study, Thermus thermophilus protein S1, which is composed only of R1-R5 domains, is not required at the early stages of trans-translation in vitro. |
| Association of protein S1 with tmRNA bound to ribosomes |
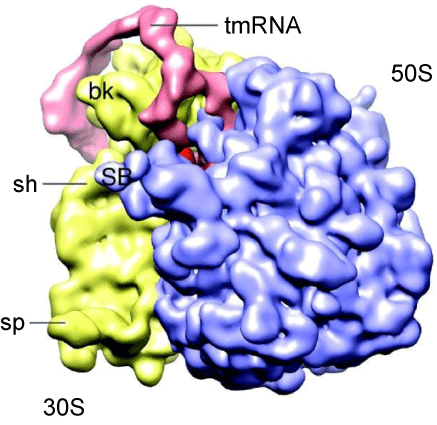
| Since intact tmRNA and protein S1 molecules have so far resisted crystallization, our insights into their three-dimensional structures and the architecture of the tmRNA:ribosome and protein S1:ribosome complexes are derived primarily from cryo-Electron Microscopy (cryo-EM) studies. The first cryo-EM structures represented the pre-accommodation and accommodation complexes of the ribosome-bound tmRNA [28-31]. At these early stages of trans-translation, the TLD enters the ribosomal A site, whereas the segment consisting of pk2, pk3 and pk4 forms an arc that remains outside the ribosome. According to Valle et al. [30] and Gillet et al. [32], the presence of protein S1, which remains invisible in the cryo-EM images, directly affects the conformation of tmRNA in the pre-accommodation complexes formed in vitro. One can argue that the observed conformational differences could not be induced by ribosome-bound protein S1, because its binding site is located at the junction of the head and the platform on the solvent side of the 30S subunit, far away from the well-established tmRNA binding site [33]. |
| In order to study the structure of tmRNA:ribosome complexes in the advanced stages of trans-translation, we prepared mutant E. coli tmRNAs, denoted in figure 1B as MS2-tmRNA-4 and MS2-tmRNA-7. As shown in figure 1B, these mutant tmRNAs have a short MLR that encodes either three or six amino acid-long peptide tags. Moreover, these mutant tmRNAs are equipped with an MS2 protein-binding hairpin inserted in helix 5. We purified MS2-tmRNA:ribosome complexes from the cell by affinity chromatography, using GST-MS2 fusion protein and Glutatione-Sepharose (Amersham Biosciences). Our analysis of the in vivo assembled complexes demonstrated that in the MS2-tmRNA-4:ribosome complex corresponded to the “resume complex”, as its A and P sites were occupied by tRNAAla (interacting with the GCA-resume codon) and TLD, respectively [34]. In contrast, in the MS2-tmRNA-7:ribosome complex, only the P site was occupied by tRNA, and the TLD was positioned outside of the ribosome. Although both tmRNA complexes were subjected to cryo-EM analysis, we were able to visualize only the resume complex (Figure 3). Our analysis demonstrated that at the resume stage of trans-translation tmRNA maintains a stable structure, which has already been observed in pre-accommodation and accommodation complexes. This observation was confirmed by cryo-EM analysis of resume complexes assembled in vitro, using T. thermophilus tmRNA and ribosomes [31]. |
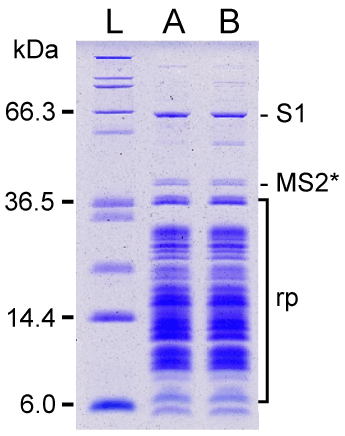
| SDS-PAGE analysis of MS2-tmRNA-4 and -7:ribosome complexes revealed that both complexes contained ribosomal protein S1 (Figure 4). Protein S1 occupies a characteristic location, far above other ribosomal proteins, on a 10% SDS-polyacrylamide gel. The identity of the protein was confirmed by MALDI-MS analysis. To estimate the quantity of protein S1 in the tmRNA:ribosome complexes, we co-separated increasing amounts of purified E. coli protein S1. Comparison of the intensities of Coomassie Blue-stained bands, corresponding to the ribosome-bound protein S1 and the reference bands of purified protein S1, showed that 0.9-1.2 molecules of S1 were bound to MS2-tmRNA:ribosome complexes. Given that conventional ribosome preparations contain less-than-stoichiometric amounts of protein S1 [35], because there is not enough protein S1 to bind to all ribosomes in logarithmically growing E. coli cells, and because ribosome-bound protein S1 readily separates from ribosomes during their preparation, our observation suggests that protein S1 is directly and strongly attached to tmRNA forming a complex composed of equimolar amounts of tmRNA and protein S1. This suggestion is also supported by strong binding of protein S1 to tmRNA:ribosome complexes in a low salt buffer, described by Tal et al. [36] (data not shown). In our earlier in vitro studies, the value of K’a for the E. coli protein S1:tmRNA interaction was determined to be ~1×108 M-1 [11]. |
| Mapping the interactions between protein S1 and tmRNA |
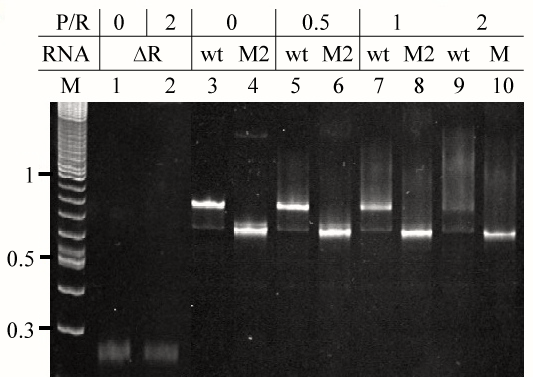
| In order to identify segments of E. coli tmRNA that constitute protein S1 binding site, we produced fourteen tmRNA mutants lacking all possible combinations of MLR, pk2, pk3 and pk4 using PCR site-directed mutagenesis [17]. We used T7 RNA polymerase-mediated in vitro run-off transcription to synthesize the mutant tmRNAs, denoted M1-M14 in table 1. Interactions between the mutant tmRNAs and intact E. coli protein S1 were analyzed using gel mobility shift assay. An example of such analysis is shown in figure 5. Binding activities of protein S1 to the intact tmRNA and the tmRNA mutants were calculated by monitoring either the formation of protein:RNA complexes or the disappearance of free RNA. Both methods produced similar results that are compiled in table 1. These data show that the protein S1 is likely to interact with a large region of tmRNA, including the MLR, pk2 and pk3, and that pk4 contributes little to the binding of protein S1. |
| Recently, we demonstrated that tmRNA lacking pseudoknot pk1 tags efficiently truncated proteins both in vitro and in vivo [37]. This observation suggests that pk1 is not a part of the protein S1 binding site on E. coli tmRNA. We used gel mobility shift assay to see whether protein S1 binds to the pk1 transcript. As control, we used a transcript that corresponded to mutant pk1, which does not fold into a pseudoknot, but is present in the tagging-competent tmRNA Δh4/Δ1p [37]. Figure 6 shows that protein S1 binds readily to the mostly single-stranded mutant pk1, but it is not able to bind to the wild-type pk1. These data are consistent with our earlier observation that a truncated tmRNA lacking MLR and pk1-pk4 cannot compete with full-length tmRNA [38]. These data are also consistent with a pattern of cross-links generated by UV irradiation of complexes formed by binding protein S1 to free and ribosome-bound photoreactive tmRNA derivatives, in which U residues are substituted by 4-thio-uridine (s4U). These cross-linked nucleotides are located exclusively in MLR (U85 and U105), pk2 (U172) and pk3 (U198, U212, U230 and U246) [11]. |
| Conclusions |
| A picture of how protein S1 is bound to tmRNA has emerged from the data collected over the past ten years. Although some in vitro studies have found that at least two protein S1 molecules could bind to E. coli tmRNA, such a complex most likely represents an artifact of the particular in vitro conditions that were applied because excess of protein S1 has been shown to inhibit protein tagging [39]. We have shown here that one molecule of protein S1 assists one tmRNA molecule, as it transits the ribosome during trans-translation (Figure 7), and the cytoplasm protein S1 binds to the tmRNA:SmpB complex. This interaction is critical at least in some bacteria, as its inhibition by the drug pyramizide aborts trans-translation in M. tuberculosis [16]. The region encompassing MLR, pk2 and pk3 constitutes the protein S1 binding site on the tmRNA molecule. The interactions between protein S1 and tmRNA are only slightly affected by tmRNA binding to the ribosome, as indicated by photoaffinity labeling experiments [11]. Recently, Qu et al. [40] demonstrated that one molecule of S1 binds to an RNA segment composed of 10 nucleotides. Since approximately one hundred nucleotides form the region consisting of MLR, pk2 and pk3, one can propose a “rolling mechanism” for protein S1 functions on tmRNA, in which protein S1 binds first to a single-stranded segment of tmRNA (e.g. MLR), and then “unzips” its structured segments (helix 5, pk2 and pk3). Such a “rolling mechanism” is supported by experiments in which optical tweezers were used to demonstrate unzipping and re-zipping of an RNA hairpin by a single protein S1 molecule in multiple steps [40]. Because the unzipping and re-zipping of double-stranded RNA segments occurs sequentially, the rolling mechanism may explain why only small local conformational changes in tmRNA can be observed, when the cryo-EM snapshots of tmRNA:ribosome complexes formed either in the presence or in the absence of protein S1 are analyzed [30,32]. Sequential rearrangements may also explain the finding that R2 of E. coli protein S1 is important for the initial binding of S1 to tmRNA [25]. Consistent with this, Saguy et al. [39] observed that truncated protein S1 lacking both R1 and R2 can, albeit inefficiently, trigger protein tagging. The contribution of R6 to protein S1 functions needs to be investigated further. Although R6 binds RNAs in vitro, it is not necessary for protein S1 functions in canonical translation and transcriptional cycling, it is dispensable for protein S1 activity in Qβ bacteriophage replication, and it is not required for protein tagging in M. tuberculosis [16,23,41,42]. |
| Our reassessment herein firmly establishes the importance of protein S1 in trans-translation. We hope that this will stimulate a new wave of interest in using this important bacterial protein as a target for pharmacological interventions in diseases caused by a substantial number of bacterial species. |
| Acknowledgements |
| We acknowledge the NIH (Grant GM58267) and NSF (Grant 1063536) for funding this project. Publication costs were supported in part by the Upchurch Fund for Excellence. |
References
|
Tables and Figures at a glance
| Table 1 |
Figures at a glance
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
 |
 |
 |
|
| Figure 5 | Figure 6 | Figure 7 |
Relevant Topics
- Analytical Biochemistry
- Applied Biochemistry
- Carbohydrate Biochemistry
- Cellular Biochemistry
- Clinical_Biochemistry
- Comparative Biochemistry
- Environmental Biochemistry
- Forensic Biochemistry
- Lipid Biochemistry
- Medical_Biochemistry
- Metabolomics
- Nutritional Biochemistry
- Pesticide Biochemistry
- Process Biochemistry
- Protein_Biochemistry
- Single-Cell Biochemistry
- Soil_Biochemistry
Recommended Journals
- Biosensor Journals
- Cellular Biology Journal
- Journal of Biochemistry and Microbial Toxicology
- Journal of Biochemistry and Cell Biology
- Journal of Biological and Medical Sciences
- Journal of Cell Biology & Immunology
- Journal of Cellular and Molecular Pharmacology
- Journal of Chemical Biology & Therapeutics
- Journal of Phytochemicistry And Biochemistry
Article Tools
Article Usage
- Total views: 14625
- [From(publication date):
specialissue-2013 - Jul 06, 2025] - Breakdown by view type
- HTML page views : 10043
- PDF downloads : 4582
