Mini Review Open Access
Quantitative Immunoproteomics Approach for the Development of MHC Class I Associated Peptide Antigens of Alpha-Cobra Toxin from Naja kaouthia
Sherkhane AS, Changbhale SS and Gomase VS*The Global Open University, Nagaland, India
- Corresponding Author:
- Virendra S Gomase
The Global Open University
Nagaland, India
E-mail: gomase.viren@gmail.com
Received date: October 30, 2014; Accepted date: November 25, 2014; Published date: December 03, 2014
Citation: Sherkhane AS, Changbhale SS, Gomase VS (2014) Quantitative Immunoproteomics Approach for the Development of MHC Class I Associated Peptide Antigens of Alpha-Cobra Toxin from Naja kaouthia. J Biotechnol Biomater 4:169. doi:10.4172/2155-952X.1000169
Copyright: © 2014 Sherkhane AS, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
Alpha-Cobratoxin from N. kaouthia binds to acetylcholine receptors which are located in neuromuscular junctions, once activated; they cause contract muscles and block their actions thereby bringing on muscle paralysis. Peptide fragments of Alpha-Cobratoxin from N. kaouthia having 71 amino acids, which shows 63 nonamers and are used synthetic peptide vaccine design and to increase the understanding of roles of the immune system against snake bite. For the immune responses against a protein antigen, it is clear that the whole protein is not necessary for raising the immune response, but small segments (15-PNGHVCYTKT-24, 26-CDAFCSIRG-34 and 36-RVDLGCAATCPTVKTGVDIQCCSTD-60) of Alpha-Cobratoxin protein from N. kaouthia called the antigenic determinants or the epitopes are sufficient for eliciting the desired immune response. The identification of specific peptides that binds to MHC class I molecules is important to recognize T-cell epitopes. In this research work, we predict antigenicity, Solvent accessibility to identify the membrane-spanning regions (hydrophobic) or regions that are likely exposed on the surface of proteins (hydrophilic domains) which are potentially antigenic that are used to design synthetic peptide vaccine.
Keywords
Alpha-cobratoxin; N. kaouthia; Antigenic peptides; MHC-binders; TapPred; PSSM
Introduction
The N. kaouthia is also called monocled cobra, is widespread across central and southern Asia regions [1]. It can adapt habitats from natural to anthropogenically impacted environments and are most active at dusk. Alpha-Cobratoxin from N. kaouthia venoms are postsynaptic neurotoxins, that have high affinity to muscular, Torpedo and neuronal alpha-7 nicotinic acetylcholine receptors which block the nerve transmission by binding specifically to the nicotinic acetylcholine receptor, leading paralysis and even death occurred due to respiratory failure [2-4]. Alpha-Cobratoxin N. kaouthia antigenic peptides are most suitable for vaccine development because an ample immune response can be generated even with single epitope. Major histocompatibility complex (MHC) molecules are cell surface proteins that binds to the peptides of antigenic proteins, present them at the cell surface and are recognized by T-cells [5,6]. T cell recognition is a fundamental mechanism by which the host identifies and responds to foreign antigens [7,8]. The MHC molecule is extremely polymorphic [9]. MHC class I molecules are expressed on most nucleated cells, generally present peptides from intracellular proteins that are targeted by proteasome, cleaved them into short peptides of 8-11 amino acids in length. These peptides are bound by the transmembrane peptide transporter (TAP) and translocate them from cytoplasm to endoplasmic reticulum, where they are bound by MHC molecule to elicit an immune response via T-cell. T cells also recognize selfpeptides but are eliminated during the thymic selection; therefore, the primary targets of T cell to recognize [10] foreign peptides and kill host target cells. The second and the C-terminal position of the peptide are the most important for binding [11,12] and the amino acids at each position contribute a certain binding energy [13]. Therefore, the identification of MHC-binding peptides and T-cell epitopes and study of antigenic properties helps to improve our understanding of specificity of immune responses are important for the development of new vaccines. However, this theme is implemented in designing synthetic peptide vaccines.
Materials and Methods
Antigenic epitopes of Alpha-Cobratoxin from N. kaouthia are determined by using the Hopp and Woods, Welling, Parker, Bepipred, Kolaskar and Tongaonkar antigenicity methods to predicts those segments which are likely to be antigenic by eliciting an immune response. [16-20]. The MHC peptide binding of antigen protein is predicted by using neural networks trained on C terminals of known epitopes. Rankpep predicts peptide binders to MHC-I ligands whose C-terminal end is likely to be the result of proteosomal cleavage using Position Specific Scoring Matrices (PSSMs) [21-28]. We predict cascade SVM based several TAP binders which was based on the sequence and the features of amino acids [29]. We also predict solvent accessible regions of proteins having highest probability that a given protein region lies on the surface of a protein Surface Accessibility, backbone or chain flexibility by Emani et al. [30] and Karplus and Schulz to identify active site of functionally important residues in membrane proteins. [31]. By using different scale we predict the hydrophobic and hydrophilic characteristics of amino acids that are rich in charged and polar residues i.e. Sweet et al., Kyte and Doolittle, Abraham and Leo, Bull and Breese, Miyazawa et al., Roseman, Wolfenden et al., Wilson et al., Cowan, Chothia [32-41].
Results and Interpretation
A antigenic sequence of Alpha-Cobratoxin from N. kaouthia is 71 residues long as- >gi| 128930|sp|P01391.1| NXL1_KAIRCFITP DITSKDCPNGHVCYTKTWCDAFCSIRGKRVDLGCAATCPTVKTGVDIQCCSTDNCNPFPTRKRP
Prediction of antigenic peptides
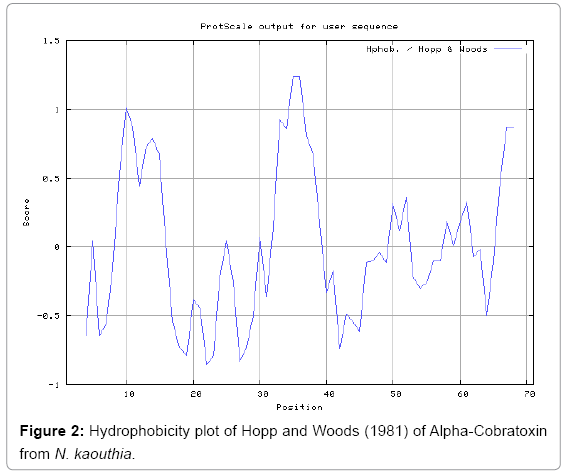
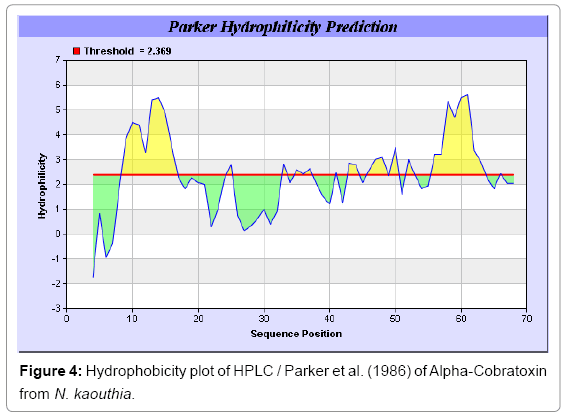
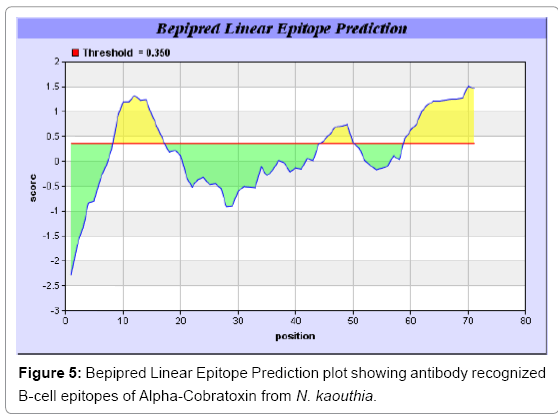
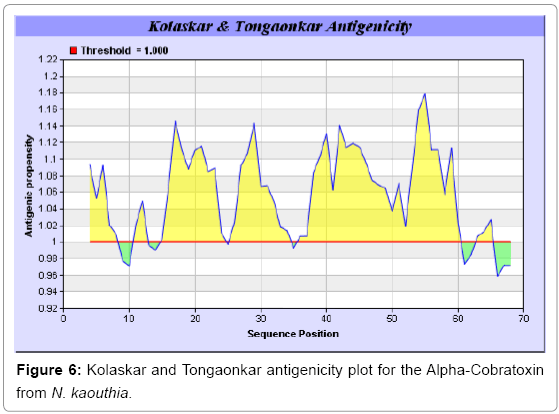
Antigenicity is predicted by identifying antigenic determinants by finding the area of greatest local hydrophilicity. The Hopp-Woods scale Hydrophilicity Prediction Result Data found high in position 9-11, 13- 15, 33-37 (1.243) in a protein, assuming that the antigenic determinants would be exposed on the surface of the protein and thus would be located in hydrophilic regions (Figures 1 and 2). Welling antigenicity plot gives value as the log of the quotient between percentage in a sample of known antigenic regions and percentage in average proteins and Prediction Result Data found high in position 20-21 (0.440), 36-38 (Figure 3). We also study Hydrophobicity plot of HPLC/Parker Hydrophilicity Prediction Result Data found 7-PDITSKD-13 (4.500), 8-DITSKDC-14 (4.400), 10-TSKDCPN-16 (5.414), 11-SKDCPNG-17 (5.486), 55-QCCSTDN- 61(5.357),57-CSTDNCN-63(5.500),58-STDNCNP-64(5.600) (maximum) (Figure 4), BepiPred predicts the location of linear B-cell epitopes Result found that 9-ITSKDCPNG-17,45-CPTVKT-50, (Figure 5) (Table 3), Kolaskar and Tongaonkara semi-empirical method used for prediction of antigenic determinants on protein antigens. Predicted peptides result found i.e. 15-PNGHVCYTKT-24, 26-CDAFCSIRG-34, 36-RVDLGCAATCPTVKTGVDIQCCSTD-60, (Figure 6) (Table 4). The maximal hydrophilicity region is assumed to be an antigenic site, having hydrophobic characteristics, because terminal regions of antigen protein are solvent accessible and unstructured; antibodies against those regions are also likely to recognize the native protein.
Solvent accessible regions
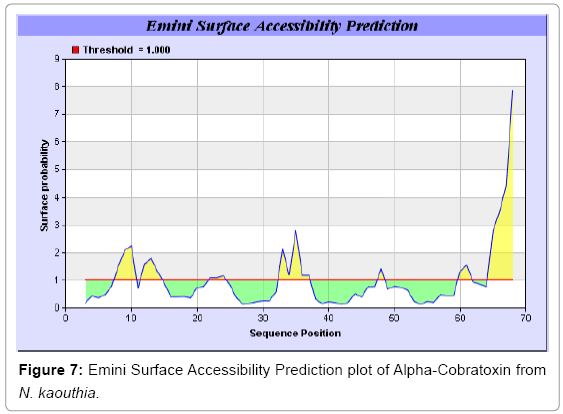
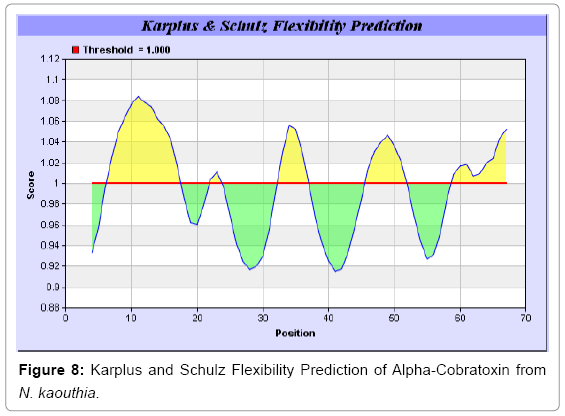
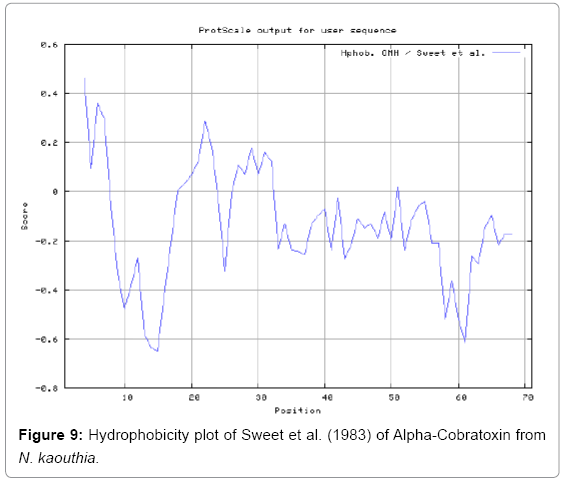
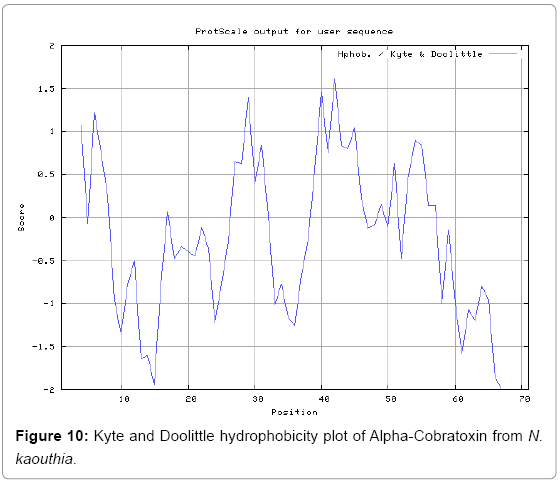
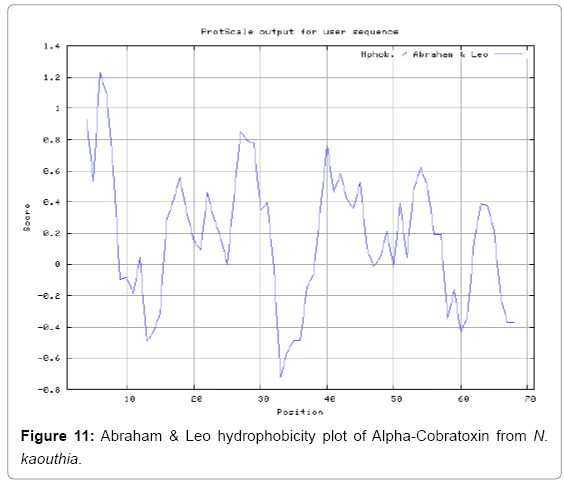
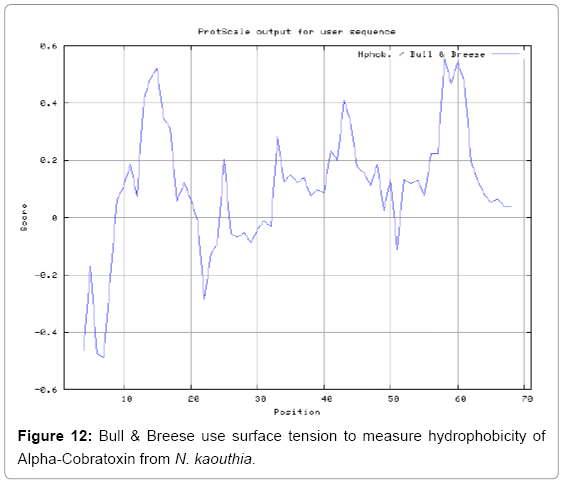
We also predict solvent accessible regions in proteins to identify antigenic activity, surface region of peptides. Emani et al., (Figure 7) predicts the highest probability i.e. found 7-PDITSK-12 (2.082), 8-DITSKD-13 (2.248), 31-SIRGKR-36 (2.121), 33-RGKRVD-38 (2.798), 63-NPFPTR-68 (2.798), 64-PFPTRK-69 (3.480), 65-FPTRKR-70 (4.408), 66-PTRKRP-71 (7.872) (maximum), that a given protein region lies on the surface of a protein and are used to identify antigenic determinants on the surface of proteins. Karplus and Schulz (Figure 8) method used for the Selection of Peptide Antigens. High score is found in residue i.e. 6-TPDITSK-12 (1.064), 7-PDITSKD-13 (1.077), 8-DITSKDC-14 (1.084) (maximum), 9-ITSKDCP-15 (1.078), 10-TSKDCPN-16 (1.074), 11-SKDCPNG-17 (1.062). Karplus and Schulz Predict backbone or chain flexibility on the basis of the known temperature B factors of the a-carbons. The hydrophobicity and hydrophilic characteristics of amino acids is determined by using different scales that are rich in charged and polar residues i.e. Sweet et al. hydrophobicity prediction Result Data found high in position 4 (0.461), 6-7, 21-23, Kyteand Doolittle result high in position 4,6-7, 29- 31, 40-44 (1.614), Abraham and Leo result data shows high in position 6-7 (1.230), 27-29, 40-42, Bull and Breese result high in position 13-16, 43-44, 58-61 (0.557),Guy result high in position 9-10, 13-15, 33-37, 66-68 (0.661), Miyazawa result high in position 4-7 (6.737), 27-31, 41- 42, 54-57, Roseman result high in position 6-7 (0.334), 17-18,42-45, Wolfenden result high in position 42 (0.170), Wilson et al. 4-6, 17-19, 22-23, 26-32 (3.671), 39-40, 54-55, Cowan 4-7 (0.899), 27-29, 40-42, Chothia4-8, 27-32 (0.407), 39-45, 53-57, (Figures 9-19).
Prediction of MHC binding peptide
We predict the peptide binders of Alpha-Cobratoxin from N. kaouthia to MHC-I molecules to a number of different alleles using Position Specific Scoring Matrix. Alpha-Cobratoxin from N. kaouthia sequence is 71 residues long, having 63 nonamers. MHC molecules are cell surface proteins, which actively participate in host immune reactions and involvement of MHC-I in response to almost all antigens. We have predicted MHC-I peptide binders of Alpha-Cobratoxin from N. kaouthia was tested with on a set of 4 different alleles i.e. H2-Db (mouse) 8mer, H2-Db (mouse) 9mer, H2-Db (mouse) 10mer, H2-Db (mouse) 11mer (Tables 1a-1d). Here RANKPEP report PSSM-specific binding threshold and is obtained by scoring all the antigenic peptide sequences included in the alignment from which a profile is derived, and is defined as the score value that includes 85% of the peptides within the set. Peptides whose score is above the binding threshold will highlighted in (Tables 1a-1d) and peptides produced by the cleavage prediction model are highlighted in (Table 2). We also use a cascade SVM based TAPP red method which found 25 High affinity TAP Transporter peptide regions which represents predicted TAP binders residues which occur at N and C termini from Alpha-Cobratoxin from N. kaouthia.
| MHC-I Allele | POS | N | SEQUENCE | C | MW (Da) | SCORE | % OPT. |
|---|---|---|---|---|---|---|---|
| 8mer_H2_Db | 41 | DLG | CAATCPTV | KTG | 746.89 | 21.518 | 40.99 % |
| 8mer_H2_Db | 61 | STD | NCNPFPTR | KRP | 930.05 | 10.742 | 20.46 % |
| 8mer_H2_Db | 58 | QCC | STDNCNPF | PTR | 878.91 | 3.74 | 7.12 % |
| 8mer_H2_Db | 2 | I | RCFITPDI | TSK | 946.14 | 0.021 | 0.04 % |
| 8mer_H2_Db | 14 | SKD | CPNGHVCY | TKT | 874.0 | -0.041 | -0.08 % |
| 8mer_H2_Db | 23 | CYT | KTWCDAFC | SIR | 932.11 | -0.625 | -1.19 % |
| 8mer_H2_Db | 18 | PNG | HVCYTKTW | CDA | 996.17 | -4.872 | -9.28 % |
| 8mer_H2_Db | 12 | ITS | KDCPNGHV | CYT | 850.94 | -5.222 | -9.95 % |
| 8mer_H2_Db | 13 | TSK | DCPNGHVC | YTK | 825.91 | -5.524 | -10.52 % |
| 8mer_H2_Db | 50 | TVK | TGVDIQCC | STD | 819.94 | -6.316 | -12.03 % |
| 8mer_H2_Db | 32 | FCS | IRGKRVDL | GCA | 938.14 | -8.761 | -16.69 % |
| 8mer_H2_Db | 22 | VCY | TKTWCDAF | CSI | 930.07 | -9.103 | -17.34 % |
| 8mer_H2_Db | 49 | PTV | KTGVDIQC | CST | 844.97 | -9.479 | -18.06 % |
| 8mer_H2_Db | 26 | KTW | CDAFCSIR | GKR | 896.06 | -9.479 | -18.06 % |
| 8mer_H2_Db | 62 | TDN | CNPFPTRK | RP | 944.12 | -15.508 | -29.54 % |
| 8mer_H2_Db | 28 | WCD | AFCSIRGK | RVD | 863.05 | -18.667 | -35.56 % |
| 8mer_H2_Db | 29 | CDA | FCSIRGKR | VDL | 948.16 | -28.837 | -54.93 % |
Table 1a: Peptide binders of Alpha-Cobratoxin from N. kaouthia to MHC-I molecules, having C-terminal ends are proteosomal cleavage sites, Binding potential (score) of antigenic peptide to the MHC-1 Allele i.e. 8mer_H2_Db.
| MHC-I Allele | POS | N | SEQUENCE | C | MW | SCORE | % OPT. |
|---|---|---|---|---|---|---|---|
| 9mer_H2_Db | 57 | IQC | CSTDNCNPF | PTR | 982.05 | 14.379 | 28.55 % |
| 9mer_H2_Db | 12 | ITS | KDCPNGHVC | YTK | 954.08 | 7.581 | 15.05 % |
| 9mer_H2_Db | 13 | TSK | DCPNGHVCY | TKT | 989.09 | 5.283 | 10.49 % |
| 9mer_H2_Db | 40 | VDL | GCAATCPTV | KTG | 803.94 | 2.756 | 5.47 % |
| 9mer_H2_Db | 61 | STD | NCNPFPTRK | RP | 1058.22 | 1.882 | 3.74 % |
| 9mer_H2_Db | 21 | HVC | YTKTWCDAF | CSI | 1093.25 | -0.651 | -1.29 % |
| 9mer_H2_Db | 22 | VCY | TKTWCDAFC | SIR | 1033.21 | -2.115 | -4.20 % |
| 9mer_H2_Db | 25 | TKT | WCDAFCSIR | GKR | 1059.27 | -2.581 | -5.12 % |
| 9mer_H2_Db | 49 | PTV | KTGVDIQCC | STD | 948.11 | -3.287 | -6.53 % |
| 9mer_H2_Db | 31 | AFC | SIRGKRVDL | GCA | 1025.22 | -6.633 | -13.17 % |
| 9mer_H2_Db | 17 | CPN | GHVCYTKTW | CDA | 1053.22 | -6.897 | -13.69 % |
| 9mer_H2_Db | 60 | CST | DNCNPFPTR | KRP | 1045.14 | -9.799 | -19.46 % |
| 9mer_H2_Db | 11 | DIT | SKDCPNGHV | CYT | 938.02 | -10.788 | -21.42 % |
| 9mer_H2_Db | 27 | TWC | DAFCSIRGK | RVD | 978.14 | -14.271 | -28.34 % |
| 9mer_H2_Db | 1 | - | IRCFITPDI | TSK | 1059.3 | -14.47 | -28.73 % |
| 9mer_H2_Db | 48 | CPT | VKTGVDIQC | CST | 944.1 | -20.758 | -41.22 % |
| 9mer_H2_Db | 28 | WCD | AFCSIRGKR | VDL | 1019.24 | -25.564 | -50.76 % |
Table 1b: Peptide binders of Alpha-Cobratoxin from N. kaouthia to MHC-I molecules, having C-terminal ends are proteosomal cleavage sites, the binding potential (score) of antigenic peptide to the MHC-1 Allele i.e. 9mer_H2_Db.
| MHC-I Allele | POS. | N | SEQUENCE | C | MW | SCORE | % OPT. |
|---|---|---|---|---|---|---|---|
| 10mer_H2_Db | 60 | CST | DNCNPFPTRK | RP | 1173.31 | 12.485 | 21.21 % |
| 10mer_H2_Db | 12 | ITS | KDCPNGHVCY | TKT | 1117.26 | 7.127 | 12.11 % |
| 10mer_H2_Db | 59 | CCS | TDNCNPFPTR | KRP | 1146.24 | 6.054 | 10.29 % |
| 10mer_H2_Db | 39 | RVD | LGCAATCPTV | KTG | 917.1 | 1.576 | 2.68 % |
| 10mer_H2_Db | 16 | DCP | NGHVCYTKTW | CDA | 1167.32 | -2.258 | -3.84 % |
| 10mer_H2_Db | 27 | TWC | DAFCSIRGKR | VDL | 1134.33 | -5.536 | -9.41 % |
| 10mer_H2_Db | 30 | DAF | CSIRGKRVDL | GCA | 1128.36 | -6.587 | -11.19 % |
| 10mer_H2_Db | 24 | YTK | TWCDAFCSIR | GKR | 1160.37 | -6.936 | -11.78 % |
| 10mer_H2_Db | 11 | DIT | SKDCPNGHVC | YTK | 1041.16 | -9.087 | -15.44 % |
| 10mer_H2_Db | 48 | CPT | VKTGVDIQCC | STD | 1047.24 | -11.614 | -19.73 % |
| 10mer_H2_Db | 10 | PDI | TSKDCPNGHV | CYT | 1039.12 | -11.992 | -20.37 % |
| 10mer_H2_Db | 47 | TCP | TVKTGVDIQC | CST | 1045.2 | -18.063 | -30.69 % |
| 10mer_H2_Db | 20 | GHV | CYTKTWCDAF | CSI | 1196.39 | -18.172 | -30.87 % |
| 10mer_H2_Db | 56 | DIQ | CCSTDNCNPF | PTR | 1085.19 | -21.639 | -36.76 % |
| 10mer_H2_Db | 21 | HVC | YTKTWCDAFC | SIR | 1196.39 | -25.354 | -43.08 % |
| 10mer_H2_Db | 26 | KTW | CDAFCSIRGK | RVD | 1081.28 | -27.786 | -47.21 % |
Table 1c: Peptide binders of Alpha-Cobratoxin from N. kaouthia to MHC-I molecules, having C-terminal ends are proteosomal cleavage sites, the binding potential (score) of antigenic peptide to the MHC-1 Allele i.e. 10mer_H2_Db.
| MHC-I Allele | POS | N | SEQUENCE | C | MW | SCORE | % OPT |
|---|---|---|---|---|---|---|---|
| 11mer_H2_Db | 11 | DIT | SKDCPNGHVCY | TKT | 1204.34 | 14.334 | 18.03 % |
| 11mer_H2_Db | 47 | TCP | TVKTGVDIQCC | STD | 1148.34 | -8.952 | -11.26 % |
| 11mer_H2_Db | 26 | KTW | CDAFCSIRGKR | VDL | 1237.47 | -9.5 | -11.95 % |
| 11mer_H2_Db | 15 | KDC | PNGHVCYTKTW | CDA | 1264.44 | -9.844 | -12.38 % |
| 11mer_H2_Db | 29 | CDA | FCSIRGKRVDL | GCA | 1275.54 | -14.187 | -17.85 % |
| 11mer_H2_Db | 59 | CCS | TDNCNPFPTRK | RP | 1274.41 | -16.292 | -20.49 % |
| 11mer_H2_Db | 20 | GHV | CYTKTWCDAFC | SIR | 1299.53 | -17.308 | -21.77 % |
| 11mer_H2_Db | 19 | NGH | VCYTKTWCDAF | CSI | 1295.52 | -18.359 | -23.09 % |
| 11mer_H2_Db | 10 | PDI | TSKDCPNGHVC | YTK | 1142.26 | -18.948 | -23.84 % |
| 11mer_H2_Db | 23 | CYT | KTWCDAFCSIR | GKR | 1288.54 | -20.282 | -25.51 % |
| 11mer_H2_Db | 46 | ATC | PTVKTGVDIQC | CST | 1142.32 | -20.512 | -25.80 % |
| 11mer_H2_Db | 9 | TPD | ITSKDCPNGHV | CYT | 1152.28 | -23.253 | -29.25 % |
| 11mer_H2_Db | 55 | VDI | QCCSTDNCNPF | PTR | 1213.32 | -24.834 | -31.24 % |
| 11mer_H2_Db | 58 | QCC | STDNCNPFPTR | KRP | 1233.32 | -24.917 | -31.34 % |
| 11mer_H2_Db | 38 | KRV | DLGCAATCPTV | KTG | 1032.19 | -25.364 | -31.91 % |
| 11mer_H2_Db | 4 | IRC | FITPDITSKDC | PNG | 1221.39 | -28.415 | -35.74 % |
| 11mer_H2_Db | 25 | TKT | WCDAFCSIRGK | RVD | 1244.49 | -30.115 | -37.88 % |
Table 1d: Peptide binders of Alpha-Cobratoxin from N. kaouthia to MHC-I molecules, having C-terminal ends are proteosomal cleavage sites the binding potential (score) of antigenic peptide to the MHC-1 Allele i.e. 11mer_H2_Db.
| Peptide Rank | Start Position | Sequence | Score | Predicted Affinity |
|---|---|---|---|---|
| 1 | 41 | CAATCPTVK | 8.618 | High |
| 2 | 30 | CSIRGKRVD | 8.586 | High |
| 3 | 38 | DLGCAATCP | 8.549 | High |
| 4 | 20 | CYTKTWCDA | 8.487 | High |
| 5 | 50 | TGVDIQCCS | 8.385 | High |
| 6 | 58 | STDNCNPFP | 8.241 | High |
| 7 | 7 | PDITSKDCP | 8.071 | High |
| 8 | 40 | GCAATCPTV | 8.010 | High |
| 9 | 32 | IRGKRVDLG | 7.958 | High |
| 10 | 55 | QCCSTDNCN | 7.872 | High |
| 11 | 36 | RVDLGCAAT | 7.814 | High |
| 12 | 62 | CNPFPTRKR | 7.738 | High |
| 13 | 47 | TVKTGVDIQ | 7.576 | High |
| 14 | 46 | PTVKTGVDI | 7.561 | High |
| 15 | 22 | TKTWCDAFC | 7.380 | High |
| 16 | 48 | VKTGVDIQC | 7.319 | High |
| 17 | 51 | GVDIQCCST | 7.318 | High |
| 18 | 33 | RGKRVDLGC | 6.855 | High |
| 19 | 44 | TCPTVKTGV | 6.789 | High |
| 20 | 15 | PNGHVCYTK | 6.764 | High |
| 21 | 16 | NGHVCYTKT | 6.535 | High |
| 22 | 17 | GHVCYTKTW | 6.355 | High |
| 23 | 29 | FCSIRGKRV | 6.308 | High |
| 24 | 11 | SKDCPNGHV | 6.130 | High |
| 25 | 61 | NCNPFPTRK | 6.085 | High |
Table 2: Cascade SVM based High affinity TAP Binders of Alpha-Cobratoxin from N. kaouthia.
Discussion
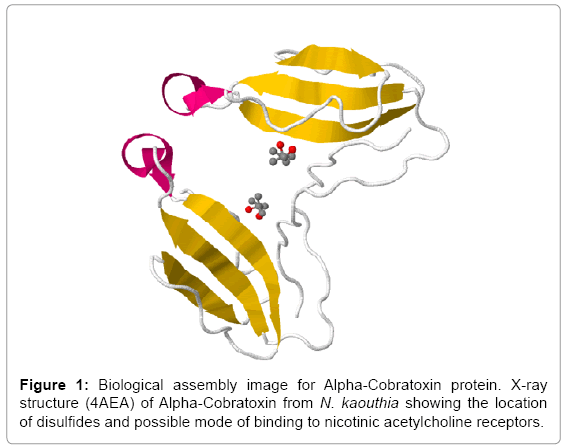
In this study, we found the antigenic determinants by finding the area of greatest local hydrophilicity. Hopp and Woods hydrophobicity scale is used to identify of potentially antigenic sites in proteins by analyzing amino acid sequences in order to find the point of greatest hydrophilic. Hydrophilicity Prediction result data found high in sequence position at 9-11, 13-15, 33-37 (1.243) in a protein this scale is basically a hydrophilic index where a polar residues have been assigned negative values. The Window size of 5-7 is good for finding hydrophilic regions, greater than 0 values are consider as hydrophilic which is consider as antigenic. Welling used information on the relative occurrence of amino acids in antigenic regions to make a scale which is useful for prediction of antigenic regions and the predicted result data found high in sequence position 20-21 (0.440), 36-38. Welling antigenicity plot gives value as the log of the quotient between percentage in a sample of known antigenic regions and percentage in average proteins. We also study Hydrophobicity plot of HPLC/Parker Hydrophilicity Prediction Result Data found 7-PDITSKD-13 (4.500), 8-DITSKDC-14 (4.400), 10-TSKDCPN-16 (5.414), 11-SKDCPNG-17 (5.486), 55-QCCSTDN-61 (5.357), 57-CSTDNCN-63 (5.500), 58-STDNCNP-64 (5.600) (maximum) (Figure 4). BepiPred predicts the location of linear B-cell epitopes Result found that there are 2 predicted epitopes are found 9-ITSKDCPNG-17, 45-CPTVKT-50, (Figure 5) (Table 3). There are 3 antigenic determinant sequences is found by Kolaskar and Tongaonkar antigenicity scales the results show highest pick at position 15-PNGHVCYTKT-24, 26-CDAFCSIRG-34, 36-RVDLGCAATCPTVKTGVDIQCCSTD-60, (Figure 6) (Table 4). Result of determined antigenic sites on proteins has revealed that the hydrophobic residues if they occur on the surface of a protein are more likely to be a part of antigenic sites. This method can predict antigenic determinants with about 75% accuracy and also gives the information of surface accessibility and flexibility. Further this region form beta sheet which show high antigenic response than helical region of this peptide and shows highly antigenicity. X-Ray Diffraction with Resolution 1.94 Å 3D Structure of the Alpha-Cobratoxin from N. kaouthia is predicted by PDB vive. We generate a purified protein for analysis of the chosen target and then structure determined the target experimentally to evaluate their similarity to known protein structures and to determine possible relationships that are identifiable from protein sequence alone. The target structure will also serve as a detailed model for determining the structure of peptide within that protein structure. We predict Solvent accessibility by using Emani et al., the result found the highest probability i.e. found 7-PDITSK-12(2.082),8- DITSKD-13 (2.248), 31-SIRGKR-36 (2.121), 33-RGKRVD-38 (2.798), 63-NPFPTR-68 (2.798), 64-PFPTRK-69 (3.480), 65-FPTRKR-70 (4.408), 66-PTRKRP-71 (7.872) (maximum), that a given protein region lies on the surface of a protein and are used to identify antigenic determinants on the surface of proteins. This algorithm also used to identify the antigenic determinants on the surface of proteins and Karplus and Schulz predict backbone or chain flexibility on the basis of the known temperature B factors of the a-carbons here we found the result with High score is i.e. 6-TPDITSK-12 (1.064), 7-PDITSKD-13 (1.077), 8-DITSKDC-14 (1.084) (maximum), 9-ITSKDCP-15 (1.078), 10-TSKDCPN-16 (1.074), 11-SKDCPNG-17 (1.062). We predict Solvent accessibility of Alpha-Cobratoxin from N. kaouthia for delineating hydrophobic and hydrophilic characteristics of amino acids. Solvent accessibility used to identify active site of functionally important residues in membrane proteins. Solvent-accessible surface areas and backbone angles are continuously varying because proteins can move freely in a three-dimensional space. The mobility of protein segments which are located on the surface of a protein due to an entropic energy potential and which seem to correlate well with known antigenic determinants. We also found the i.e. Sweet et al. hydrophobicity prediction result data found high in position 4 (0.461), 6-7, 21-23, Kyte and Doolittle result high in position 4, 6-7, 29-31, 40-44 (1.614), Abraham and Leo result high in position 6-7(1.230), 27-29, 40-42, Bull and Breese result high in position 13-16, 43-44, 58-61 (0.557), Guy result high in position 9-10, 13-15, 33-37, 66-68 (0.661), Miyazawa result high in position 4-7 (6.737), 27-31, 41-42, 54-57, Roseman result high in position 6-7 (0.334), 17-18, 42-45, Wolfenden result high in position 42 (0.170), Wilson et al. 4-6, 17-19, 22-23, 26-32 (3.671), 39- 40, 54-55, Cowan 4-7(0.899), 27-29, 40-42, Chothia 4-8,27-32 (0.407), 39-45, 53-57, (Figures 9-19). These scales are a hydrophilic with a polar residues assigned negative value. Because the N- and C-terminal regions of proteins are usually solvent accessible and unstructured, antibodies against those regions recognize the antigenic protein. In this study, we found predicted MHC-I peptide binders of toxin protein for 8mer_H2_Db alleles with the consensus sequence QNWNCCTI that yields the maximum score i.e. 52.494, 9mer_H2_Db with, the consensus sequence FCIHNCDYM that yields the maximum score i.e. 50.365, 10mer_H2_Db with, the consensus sequence SGYYNFFWCL that yields the maximum score i.e. 58.858, 11mer_H2_Db with, the consensus sequence CGVYNFYYCCY that yields the maximum score i.e. 79.495. We also use a cascade SVM based TAPP red method which found 25 High affinity TAP Transporter peptide regions which represents predicted TAP binders residues which occur at N and C termini from Alpha-Cobratoxin from N. kaouthia. TAP is an important transporter that transports antigenic peptides from cytosol to ER. TAP binds and translocate selective antigenic peptides for binding to specific MHC molecules. The efficiency of TAP-mediated translocation of antigenic peptides is directly proportional to its TAP binding affinity. Thus, by understanding the nature of peptides, that bind to TAP with high affinity, is important steps in endogenous antigen processing. The correlation coefficient of 0.88 was obtained by using jackknife validation test. In this test, we found the MHCI and MHCII binding regions. T cell immune responses are derived by antigenic epitopes hence their identification is important for design synthetic peptide vaccine. T cell epitopes are recognized by MHCI molecules producing a strong defensive immune response against Alpha-Cobratoxin from N. kaouthia. Therefore, the prediction of peptide binding to MHCI molecules by appropriate processing of antigen peptides occurs by their binding to the relevant MHC molecules. Because, the C-terminus of MHCI-restricted epitopes results from cleavage by the proteasome and thus, proteasome specifity is important for determine T-cell epitopes. Consequently, RANKPEP also focus on the prediction of conserved epitopes. C-terminus of MHCI-restricted peptides is generated by the proteasome, and thus RANKPEP also determines whether the C-terminus of the predicted MHCI-peptide binders is the result of proteasomal cleavage. Moreover, these sequences are highlighted in purple in the output results. Proteasomal cleavage predictions are carried out using three optional models obtained applying statistical language models to a set of known epitopes restricted by human MHCI molecules as indicated here.
| No. | Start Position | End Position | Peptide | Peptide Length |
|---|---|---|---|---|
| 1 | 9 | 17 | ITSKDCPNG | 9 |
| 2 | 45 | 50 | CPTVKT | 6 |
Table 3: Predicted Antigenic epitopes of Alpha-Cobratoxin from N. kaouthia Bepipred.
| No. | Start Position | End Position | Peptide | Peptide Length |
|---|---|---|---|---|
| 1 | 15 | 24 | PNGHVCYTKT | 10 |
| 2 | 26 | 34 | CDAFCSIRG | 9 |
| 3 | 36 | 60 | RVDLGCAATCPTVKTGVDIQCCSTD | 25 |
Table 4: Predicted Antigenic epitopes of Alpha-Cobratoxin from N. kaouthia.
Conclusion
From the above result and discussion it is concluded that RANKPEP predict Peptide binders of Alpha-Cobratoxin from N. kaouthia to MHC-I molecules and thereby potential T-cell epitopes. The specificity of transporter associated with antigen processing (TAP) plays an important role in the transport of the antigenic peptide fragments of the proteolysed to the endoplasmic reticulum where they associate with the major histocompatibility complex (MHC) class I molecules. Therefore, prediction of TAP-binding peptides is highly helpful in identifying the MHC class I-restricted T-cell epitopes and hence useful in the synthetic peptide vaccine designing. All above prediction methods are based on propensity scales for the 20 amino acids to describe the tendency of each residue to be associated with properties such as hydrophilicity, surface accessibility or mobility. Antigenic peptides should be located in solvent accessible regions containing both hydrophobic and hydrophilic residues. High peaks in the surface accessibility plot predict regions that have a higher chance of producing antibodies that can bind to native protein. This means the increase in affinity of MHC binding peptides may result in enhancement of immunogenicity of Alpha-Cobratoxin from N. kaouthia and are helpful in the designing of synthetic peptide vaccine. This approach can help reduce the time and cost of experimentation for determining functional properties of Alpha-Cobratoxin from N. kaouthia. Overall, the results are encouraging; both the sites of action and physiological functions can be predicted with very high accuracies helping minimize the number of validation experiments.
References
- Bain RH (2009) Najakaouthia. IUCN Red List of Threatened Species. Version 2011.1. International Union for Conservation of Nature.
- Martin BM, Chibber BA, Maelicke A (1983) The sites of neurotoxicity in alpha-cobratoxin. J Biol Chem 258: 8714-8722.
- Servent D, Winckler-Dietrich V, Hu HY, Kessler P, Drevet P, et al. (1997) Only snake curaremimetic toxins with a fifth disulfide bond have high affinity for the neuronal alpha7 nicotinic receptor. J Biol Chem 272: 24279-24286.
- Antil S, Servent D, Menez A (1999) Variability among the sites by which curaremimetic toxins bind to torpedo acetylcholine receptor, as revealed by identification of the functional residues of alpha-cobratoxin. J. Biol. Chem 274: 34851-34858.
- Rammensee HG1, Falk K, Rötzschke O (1993) Peptides naturally presented by MHC class I molecules. Annu Rev Immunol 11: 213-244.
- Cresswell P (1994) Assembly, transport, and function of MHC class II molecules. Annu Rev Immunol 12: 259-293.
- Batalia MA, Collins EJ (1997) Peptide binding by class I and class II MHC molecules. Biopolymers 43: 281-302.
- Flower DR (2008) Vaccines: how they work, in Bioinformatics for Vaccinology. Wiley-Blackwell, Oxford, UK 73–112.
- Williams TM (2001) Human leukocyte antigen gene polymorphism and the histocompatibility laboratory. J Mol Diagn 3: 98-104.
- von Boehmer H (1991) Positive and negative selection of the alpha beta T-cell repertoire in vivo. Curr Opin Immunol 3: 210-215.
- Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanović S (1999) SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50: 213-219.
- Falk K, Rötzschke O, Rammensee HG (1990) Cellular peptide composition governed by major histocompatibility complex class I molecules. Nature 348: 248-251.
- Stryhn A, Pedersen LO, Romme T, Holm CB, Holm A, et al. (1996) Peptide binding specificity of major histocompatibility complex class I resolved into an array of apparently independent subspecificities: quantitation by peptide libraries and improved prediction of binding. Eur J Immunol 26: 1911-1918.
- Dadley-Moore DL, Lightowlers MW, Rothel JS, Jackson DC (1999) Synthetic peptide antigens induce antibodies to Taenia ovis oncospheres. Vaccine 17: 1506-1515.
- McDonald DL, Stockwin T, Matzow ME Blair Zajdel GE Blair (1999) Coxsackie and adenovirus receptor (CAR)-dependent and major histocompatibility complex (MHC) class I-independent uptake of recombinant adenoviruses into human tumour cells Gene Ther 6: 1512-1519.
- Hopp TP, Woods KR (1981) Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A 78: 3824-3828.
- Welling GW, Weijer WJ, van der Zee R, Welling-Wester S (1985) Prediction of sequential antigenic regions in proteins. FEBS Lett 188: 215-218.
- Parker JM, Guo D, Hodges RS (1986) New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry 23:25
- Larsen JE, Lund O, Nielsen M (2006) Improved method for predicting linear B-cell epitopes. Immunome Res 2: 2.
- Kolaskar AS, Tongaonkar PC (1990) A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett 276: 172-174.
- Reche PA, Glutting JP, Reinherz EL (2002) Prediction of MHC class I binding peptides using profile motifs. Hum Immunol 63: 701-709.
- Reche PA, Reinherz EL (2003) Sequence variability analysis of human class I and class II MHC molecules: functional and structural correlates of amino acid polymorphisms. J Mol Biol 331: 623-641.
- Craiu A, Akopian T, Goldberg A, Rock KL (1997) Two distinct proteolytic processes in the generation of a major histocompatibility complex class I-presented peptide. Proc Natl Acad Sci U S A 94: 10850-10855.
- Gomase VS (2006) Prediction of antigenic epitopes of neurotoxin Bmbktx1 from Mesobuthus martensii. Curr Drug Discov Technol 3: 225-229.
- Sherkhane AS, Changbhale SS, Chitlange NR, Waghmare S, Gomase VS, et al. (2012) Prediction of Major Histocompatibility Complex Binding Peptides and Epitopes from NajanajaCardiotoxin (CTX). Drug Invention Today 4: 435-438.
- Gomase VS, Kale KV, Chikhale NJ, Changbhale SS (2007) Prediction of MHC binding peptides and epitopes from alfalfa mosaic virus. Curr Drug Discov Technol 4: 117-215.
- Gomase VS, Chitlange NR, Sherkhane AS, Changbhale SS, Kale KV (2013) Prediction of WuchereriaBancrofti Troponin Antigenic Peptides: Application in Synthetic Vaccine Design to Counter Lymphatic Filariasis. J Vaccines Vaccin 4: 169.
- Gomase VS, Shyamkumar K (2009) Prediction of antigenic epitopes and MHC binders of neurotoxin alpha-KTx 3.8 from Mesobuthustamulussindicus. African Journal of Biotechnology 8: 6658-6676.
- Bhasin M, Raghava GP (2004) Analysis and prediction of affinity of TAP binding peptides using cascade SVM. Protein Sci 13: 596-607.
- Emini EA, Hughes JV, Perlow DS, Boger J (1985) Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol 55: 836-839.
- Karplus PA, Schulz GE (1985) Prediction of chain flexibility in proteins: a tool for the selection of peptide antigen. Naturwissenschaften 72: 212-213.
- Sweet RM, Eisenberg D (1983) Correlation of sequence hydrophobicities measures similarity in three-dimensional protein structure. J Mol Biol 171: 479-488.
- Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105-132.
- Abraham DJ, Leo AJ (1987) Extension of the fragment method to calculate amino acid zwitterion and side chain partition coefficients. Proteins 2: 130-152.
- Bull HB, Breese K (1974) Surface tension of amino acid solutions: a hydrophobicity scale of the amino acid residues. Arch Biochem Biophys 161: 665-670.
- Miyazawa S, Jernigen RL (1985) Estimation of Effective Interresidue Contact Energies from Protein Crystal Structures: Quasi-Chemical Approximation. Macromolecules 18: 534-552.
- Roseman MA (1988) Hydrophilicity of polar amino acid side-chains is markedly reduced by flanking peptide bonds. J Mol Biol 200: 513-522.
- Wolfenden R, Andersson L, Cullis PM, Southgate CC (1981) Affinities of amino acid side chains for solvent water. Biochemistry 20: 849-855.
- Wilson KJ, Honegger A, Stötzel RP, Hughes GJ (1981) The behaviour of peptides on reverse-phase supports during high-pressure liquid chromatography. Biochem J 199: 31-41.
- Cowan R, Whittaker RG (1990) Hydrophobicity indices for amino acid residues as determined by high-performance liquid chromatography. Pept Res 3: 75-80.
- Chothia C (1976) The nature of the accessible and buried surfaces in proteins. J Mol Biol 105: 1-12.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 14896
- [From(publication date):
December-2014 - Apr 26, 2025] - Breakdown by view type
- HTML page views : 10308
- PDF downloads : 4588