Research Article Open Access
Quantitative Analysis of Anti-Hevea brasiliensis Antibody Cross-Reactivity against Taraxacum kok-saghyz Latex Proteins Demonstrates Significantly Reduced Antibody Recognition
Julian T Dafoe, Fang Huang and Trent Chunzhong Yang*
National Research Council Canada, Aquatic and Crop Resource Development Portfolio, 100 Sussex Drive, Ottawa, Ontario, Canada K1N 5A2
- *Corresponding Author:
- Yang TC
National Research Council Canada
Aquatic and Crop Resource Development Portfolio
100 Sussex Drive, Ottawa, Ontario
Canada K1N 5A2
Tel: +1 (613) 990-2114
E-mail: trent.yang@nrc-cnrc.gc.ca
Received date: August 22, 2017; Accepted date: September 23, 2017; Published date: September 30, 2017
Citation: Dafoe JT, Huang F, Yang TC (2017) Quantitative Analysis of Anti-Hevea brasiliensis Antibody Cross-Reactivity Against Taraxacum kok-saghyz Latex Proteins Demonstrates Significantly Reduced Antibody Recognition. J Biotechnol Biomater 7:270. doi:10.4172/2155-952X.1000270
Copyright: © 2017 NRC Canada. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
The annual plant Taraxacum kok-saghyz, commonly called Russian dandelion, has recently gained commercial attention as an alternative source to the rubber tree, Hevea brasiliensis, for natural rubber latex. While producing rubber of equivalent quality, the potential application of TKS latex for hypoallergenic products remains to be determined. In order to quantify the allergenicity of TKS latex proteins, we compared the extent of anti-Hevea latex polyclonal and monoclonal antibody cross-reactivity toward Hevea latex proteins to that of TKS latex proteins by quantitative ELISA methods and semi-quantitative image densitometry. ELISA measurement of polyclonal antibody recognition toward TKS latex proteins was on the order of one-tenth relative to equal amounts of Hevea latex protein, while recognition by the monoclonal antibody was below 2%. Immunoblots confirmed significantly reduced polyclonal antibody recognition toward TKS latex proteins, in the same range as the cross-reactivity exhibited toward the rubber-producing plant, Lactuca sativa and Glycine max, a non-rubber-producing plant. Despite the presence of cross-reactivity, these quantitative results support TKS as an alternative source of latex that may require lessintensive processing for the reduction of antigenic proteins in the manufacture of hypoallergenic natural rubber latex products.
Keywords
Taraxacum; Hevea; Natural rubber; Latex; Allergy; Allergenicity; Cross-reactivity; ELISA; Hypersensitivity; Hypoallergenic
Introduction
The natural rubber latex industry currently relies almost solely on environmentally-deleterious tropical plantations of the rubber tree, Hevea brasiliensis [1,2]. Furthermore, the potential for development of sensitization toward proteins contained in Hevea-derived latex poses significant health risks in the form of type I IgE-mediated hypersensitivity (allergic) reactions, particularly with the use of gloves and other medical devices in contact with skin.
Latex is typically a white sap that is stored in laticifer cells and exuded upon injury possibly to defend against herbivores and pathogens [3,4]. The latex of many plants contains rubber (cis-1,4- isoprene polymer) which accumulates within lipid-bound microscopic rubber particles. In addition to rubber, latex contains a variety of small molecules and proteins such as proteases, chitinases and glucosidases [4]. Latex proteins can be divided into three groups: soluble proteins, proteins associated with organelles, and proteins bound to the surface of rubber particles. About one quarter of the proteins in H. brasiliensis latex are tightly bound to the hydrophobic surface of rubber particles, making them difficult to remove from the resulting natural rubber latex products [5].
Improvements to manufacturing practices through washing of natural rubber latex products derived from H. brasiliensis have resulted in products with reduced allergen contents [6]. However, to produce Hevea latex safe for use by latex-hypersensitive individuals, the complete removal of all allergenic proteins is needed. Removal of the rubber particle-bound proteins would necessitate drastic treatment e.g. with proteases [7,8] and/or detergents, that would not only be costly, but could also affect the quality of the resulting latex products [5]. Alternative sources of natural rubber latex having reduced levels of antigenicity could reduce processing costs and provide an additional margin of safety. Such materials may surpass today’s Hevea-derived hypoallergenic products, which have had their antigenicity sufficiently reduced to avoid sensitization, and may indeed be tolerable for latex-allergic individuals [6]. There is no agreed-upon safe antigen concentration limit to denote latex as being hypoallergenic. However, a lower antigenic burden in latex may reduce the potential development of sensitization in previously-insensitive individuals [9]. By measuring Hevea allergens quantitatively, it may be possible to set a threshold below which gloves containing low or insignificant amounts of allergens can be considered as hypoallergenic [10].
Taraxacum kok-saghyz (TKS), also called Russian dandelion, is a rapidly-growing annual plant with high levels of rubber accumulation in its roots. It has gained increasing attention as an alternative, domestic, crop for natural rubber latex production [11]. A recent report demonstrated the presence of anti-Hevea antibody cross-reactivity toward TKS latex proteins, and questioned the crop’s potential for use in hypoallergenic products on that basis [12]. However, this conclusion was based on qualitative results with only an approximate quantitative range given; a rigorous quantitative analysis of TKS latex protein antigenicity compared with Hevea latex proteins remains unavailable.
The objective of this study was to measure the degree of anti-Hevea antibody cross-reactivity toward TKS latex proteins using quantitative ELISA assays and semi-quantitative Western blot image densitometry to substantiate the quantitative results. For further comparison, polyclonal anti-Hevea antibody cross-reactivity to proteins from TKS latex, Lactuca sativa (lettuce), another rubber-producing food crop, and a non-rubber-producing food crop, Glycine max (soybean), was evaluated by Western blot image densitometry alongside TKS latex proteins. Furthermore, potentially cross-reactive proteins in TKS, lettuce, and soybean were identified by BLAST searches using the major Hevea latex allergens as query sequences and compared on the basis of their evolutionary relationships. These findings will help in evaluating the potential applications of TKS latex for manufacturing hypoallergenic latex products.
Materials and Methods
Unless otherwise indicated, all chemicals were purchased from Sigma-Aldrich (Oakville, Ontario, Canada) or Fisher Scientific (Mississauga, Ontario, Canada).
Latex extraction from roots
Greenhouse-grown Taraxacum kok-saghyz plants were supplied by NovaBioRubber Green Technologies, Inc. (Surrey, British Columbia, Canada) and shipped overnight on ice. Latex was extracted from roots as described [13,14]. Briefly, washed roots from ten plants were homogenized for 2 min in a chilled Waring laboratory blender at 20% (w/v) in ice-cold homogenization buffer containing protease inhibitors to avoid endogenous proteolysis (100 mM Tris-HCl, pH 7.5, 5 mM MgSO4, 1% (w/v) ascorbic acid, 50 μL Antifoam 204, 5 mM 2-mercaptoethanol, and 10 mM phenylmethylsulphonyl fluoride (PMSF)). The homogenate was filtered through four layers of cheesecloth, and the filtrate underwent repeated centrifugation with collection of the top latex layer into sample tubes kept on ice.
Extraction and quantification of latex proteins
Proteins in the extracted latex were solubilized by incubation in an equal volume of extraction buffer (50 mM Na2PO4, pH 7.4, 1% (w/v) sodium dodecyl sulphate (SDS)) for 2 h at room temperature as described [12]. The rubber phase was removed by centrifugation for 15 min at 21,000x g, the aqueous phase containing solubilized latexassociated proteins was carefully removed with an 18-gauge needle and clarified by passing through a 0.45 μm low-protein-binding syringe filter for storage in aliquots at -20°C. Proteins were also extracted from a commercial sample of ammoniated Hevea natural rubber latex (KreemTEX, Enviromolds, Salem, New Jersey) and from latex obtained directly from romaine lettuce (Lactuca sativa) using a similar procedure. Proteins were extracted from ground soybeans (Glycine max) by aqueous extraction and analyzed directly.
Protein concentrations in the latex samples were quantified to permit their comparison on an equal basis in immunological assays. To remove interference from non-protein components, 1 mL samples were treated with 0.1 mL of 1.5% (w/v) sodium deoxycholate followed by precipitation of proteins using 0.2 mL of 72% (w/v) tricholoracetic acid [15]. The precipitated proteins were pelleted by centrifugation, dried, and then dissolved in 50 mM NaOH. Protein was quantified by the bicinchoninic acid, reducing agent compatible (BCA-RAC) protein assay (Thermo Fisher Scientific) calibrated using dilutions of a bovine serum albumin analytical standard (Bio-Rad, Hercules, California, USA) prepared using identical precipitation and dissolution steps. This assay provided greater sensitivity and linearity than the modified Lowry assay recommended in ASTM method D5712 for the measurement of extractable protein from latex (https://tools.thermofisher.com/content/ sfs/brochures/1602063-Protein-Assay-Handbook.pdf) [16]. Protein concentrations in the samples were determined as: 986 ± 88 μg/mL for the Hevea standard antigen (Latex natural rubber protein standard antigen, Akron Rubber Development Laboratory (ARDL), Akron, Ohio, USA); 175 ± 48 μg/mL for the Hevea latex protein extract; 539 ± 40 μg/ mL for the Taraxacum latex protein extract; 26,995 ± 1,803 μg/mL for the romaine lettuce latex extract; 7,365 ± 226 μg/mL for the soybean extract.
Polyclonal and monoclonal ELISA
Table 1 describes the antibodies used to evaluate cross-reactivity toward latex proteins. Antigenic protein in TKS latex was quantified via polyclonal inhibition enzyme-linked immunosorbent assay (ELISA) described in ASTM D6499-12 using commercially-available polyclonal anti-Hevea rabbit serum (ARDL) [17]. This assay is based on a signal inhibition proportional to the amount a sample’s of competitive binding. To quantify anti-Hevea antibody cross-reactivity toward TKS, concentrations of TKS protein which demonstrated measurable inhibition in the ELISA assay proportional to two sequential duplicate dilutions (four measurements) were related to the concentration of standard Hevea antigen producing an equal level of inhibition. That is, the antigenicity of TKS latex could be described as a ratio of the amount of TKS protein relative to Hevea protein that produced an equal level of inhibition.
| Antibody | Target | Manufacturer | Limit of Detection for ELISA |
|---|---|---|---|
| Rabbit Polyclonal | Full protein profile of Hevea NRL | Akron Rubber Development Laboratories (Akron, USA) | 30 µg/L |
| Mouse Monoclonal | Hev b 3 protein of Hevea NRL | Icosagen AS (Tartumaa, Estonia) | 0.8 µg/L |
Table 1: Properties of antibodies used for ELISA and Western blot analyses.
Allergenic protein in TKS latex was also quantified by direct monoclonal antibody recognition of the Hevea latex small rubber particle protein (SRPP), Hev b 3, using the immunoenzymometric twolayer Hev b 3 ELISA (Icosagen AS, Tartumaa, Estonia) as described in ASTM D7247-14 [18]. An antibody targeting Hev b 3 was selected because its homolog, SRPP3, is the most abundant of the identified Hevea allergens present in TKS latex, and this antibody also showed the most intense cross-reactivity to TKS latex proteins [12,19].
All ELISA quantifications were performed at least in duplicate, experiments were repeated in triplicate. Data were analyzed by Welch’s one-way ANOVA (α=0.01) using the Real Statistics Resource Pack software (Release 5.0) by Charles Zaiontz (http://www.real-statistics. com).
SDS-PAGE and Western blots
Proteins were separated by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) using gradient gels (4-15% acrylamide, Bio-Rad) loaded with equal volumes of sample buffer for Western blot and silver staining [20]. Gels were developed for total protein visualization by silver staining, and duplicate gels were transferred to polyvinylidene fluoride membranes by a modification of the method of Towbin et al. [21]. Membranes were blocked for 1 h at room temperature with 5% (w/v) bovine serum albumin in Tris-buffered saline (pH 7.6) containing Tween-20 (0.1% v/v) (TBST) followed by three washes with TBST. Blocked membranes were incubated for 1 h at room temperature with antibodies diluted in TBST (1:2500 rabbit polyclonal (ARDL); 1:1000 mouse monoclonal (anti-Hev b 3, Icosagen AS) followed by three washes in TBST. For detection, the membranes were incubated (1:10 000) with StrepTacin-Horseradish Peroxidase conjugate (Bio-Rad) for visualization of molecular weight markers and a secondary horseradish peroxidase-conjugated antibody (1:1000) (Sigma-Aldrich) against the primary antibody species of origin. Antibody binding was detected by enhanced chemiluminescence (ECL) using SuperSignal West Dura Extended Duration substrate (Fisher Scientific) and imaged using a Bio-Rad ChemiDoc MP system (Bio-Rad). Densitometric image analysis was performed using ImageLab software v5.2 (Bio-Rad) with each lane quantified as a single band, with background subtracted using a disk size of 10 mm. Protein loadings that produced a linear response in signal intensity as a function of loading were used for comparison (data not shown).
Protein homology
BLAST searches (https://blast.ncbi.nlm.nih.gov/Blast.cgi) were conducted using the amino acid sequence for each of the 15 identified Hevea allergens (Hev b 1 through Hev b 15) as the query against proteins in each of the plant species used for comparison: T. koksaghyz, L. sativa and G. max. Search results were sorted by identity score and the top five hits for each species were tabulated after removal of redundancies to illustrate the relative abundance of proteins sharing significant homology with the Hevea antigens.
A phylogenetic tree was constructed using Phylogeny Analysis tools (http://www.phylogeny.fr/) in one-click mode to illustrate the evolutionary relationships between two Hevea antigens (Hev b 1 and Hev b 3) having the greatest number of homologous proteins with the other plant species. Their homologs identified by BLAST searches for each plant species were used for comparison [22-24].
Results
Quantitative ELISA analyses of anti-Hevea antibody crossreactivity toward TKS latex proteins
Inhibition due to binding of the polyclonal antibody to the TKS latex protein competitor was detectable, in agreement with the crossreactivity reported by Cornish et al. [12]. The concentrations of total and antigenic proteins in the Hevea and TKS latex samples are given in Table 2 along with the corresponding relative antigenicity of the total protein amounts.
| Sample (total protein concentration, µg/mL ± SD) | Equivalent antigenic protein concentration, µg/mL ± SD | Antigenicity of total protein |
|---|---|---|
| Hevea protein standard antigen (986 ± 88) | 997 ± 42a | 1.01 |
| Hevealatex protein extract (175 ± 48) | 170 ± 8b | 0.97 |
| T. kok-saghyz latex protein extract (539 ± 40) | 62 ± 2c | 0.12 |
Table 2: Antigenic protein detected by rabbit polyclonal anti-Hevea antibody inhibition ELISA assay.
Superscript letters denote significantly different results (p<0.01).
Based on the polyclonal inhibition ELISA assay, TKS latex proteins exhibited only 12% of the antigenicity of an equivalent amount of Hevea latex proteins. In contrast, antigenic protein concentrations in both the HSA and the Hevea natural rubber latex were very similar to the total protein concentration measured in each sample.
Similar analyses have been reported, albeit with greater uncertainty and variability possibly due to interference in the protein quantification assays [12].
Based on our inhibition ELISA results, ~8-fold greater amount of TKS latex protein relative to Hevea latex protein was required to produce equal levels of inhibition. The level of inhibition produced by TKS latex proteins in the polyclonal inhibition ELISA assay was below the suggested inhibition signal range of 40-60% for quantification of antigenicity using this assay [17]. The low signal level could not be improved by increasing the concentration of TKS protein in solution, giving a plateau of around 25% inhibition at sample protein concentrations from 5 to 250 μg/mL, preventing direct comparison of antigenic protein concentrations in terms of Ag50 values that produced 50% inhibition. This was likely due to reduced antibody recognition, because the Hevea latex protein sample extracted in an identical fashion did not demonstrate the same plateau at low levels of inhibition. Despite the low level of inhibition, measurements had good reproducibility in replicate assays, and could be directly compared based on concentrations producing lower levels of inhibition.
The concentrations of antigenic proteins were also measured by direct two-layer ELISA using the monoclonal anti-Hev b 3 antibody. Four monoclonal anti-Hevea antibodies are commercially available; the anti-Hev b 3 monoclonal antibody was selected for ELISA and Western blots because this antibody demonstrated the strongest crossreactive signal toward TKS latex proteins among all four commercial monoclonal antibodies [12]. While protein concentrations measured by the polyclonal inhibition EILSA assay are expressed in micrograms per millilitre, Hev b 3 equivalent concentrations are three orders of magnitude lower, expressed in the range of micrograms per litre.
| Sample (total protein concentration, µg/mL ± SD) | Hev b 3 equivalent concentration (µg/L) ± SD |
|---|---|
| Hevea latex protein standard antigen (986 ± 88) | 114 ± 31a |
| Hevealatex extract (175 ± 48) | 15 ± 4b |
| Taraxacum kok-saghyz latex protein extract (539 ± 40) | 0.8 ± 0.6c |
Table 3: Antigenic protein detected by mouse monoclonal anti-Hev b 3 antibody in two-layer ELISA assay. Superscript letters denote significantly different results (p<0.01).
The monoclonal anti-Hev b 3 antibodies recognized less than 1 μg/L equivalent Hev b 3 protein in TKS latex protein samples containing 540 μg/mL total protein, representing only approximately 2.0 × 10-4% of the total protein (Table 3). In comparison, the antigenic Hev b 3 concentration in both the Hevea latex protein samples represented approximately 0.01% of the total protein concentration for each sample. As with the polyclonal ELISA assay, the detectable concentration of antigenic protein in the TKS latex protein extract was below the limit of quantification for the assay, stated by the manufacturer as 10 μg/L equivalent Hev b 3 proteins [18]. These results indicated that the intensity of monoclonal antibody recognition of proteins in TKS latex that are homologous to Hev b 3 is less than 2% compared to an equal amount of Hevea latex proteins.
Semi-quantitative Western blot analyses of TKS latex protein cross-reactivity against Hevea antibodies
Figure 1: Total protein silver stain (A); Polyclonal anti-Hevea Western blot (B); and monoclonal anti-Hev b 3 Western blot (C) of antibody recognition toward Hevea and TKS latex proteins. All sample lanes were loaded with 0.5 µg protein.
M: Molecular Mass Marker; HSA: Hevea Standard Antigen; HL: Hevea Latex Protein Extract; TL: TKS Latex Protein Extract
TKS latex proteins and Hevea latex proteins were extracted by the same procedure and visualized by silver staining (Figure 1A). Antibody binding was compared using an equal amount of protein loaded in each lane. Recognition of TKS latex proteins (TL) by polyclonal (Figure 1B) and monoclonal (Figure 1C) antibodies was compared to that of Hevea standard antigen (HSA) and Hevea latex protein extract (HL). A negative control using bovine serum albumin (Bio-Rad) included in preliminary experiments demonstrated no antibody cross-reactivity (data not shown).
The smearing in the lanes indicated that spontaneous proteolysis likely occurred during shipment and storage of the fresh roots or during sample preparation, despite efforts to prevent hydrolysis by including protease inhibitors in the extraction buffer. However, the partially hydrolyzed samples retained their antigenicity, as seen by antibody binding shown by Western blot (Figure 1B). Hydrolysis of proteins occurs during latex production as a result of endogenous proteolysis and processing conditions, and may in fact increase the number of antigenic epitopes by exposing buried sequences [25].
Western blot imaging showed that for both antibodies, the recognition of Hevea latex proteins (HSA and HL) appears qualitatively similar based on relative lane densities. In contrast, antibody recognition of TKS latex proteins (TL) relative to Hevea proteins was significantly reduced for both antibodies.
We measured the signal intensities of each lane by semiquantitative densitometric image analysis and calculated the ratios of signal intensities (Table 4). The semi-quantitative results confirmed a significant reduction in antibody binding toward TKS latex protein. Binding intensity signals of the polyclonal antibody toward TKS latex proteins were 19% of the Hevea latex standard antigen and 14% of the Hevea latex proteins for equal amounts of protein. Monoclonal anti-Hev b 3 antibody recognition of TKS latex proteins was further reduced, producing a signal intensity that was only 3% relative to Hevea latex standard antigen and 9% relative to Hevea latex proteins extracted in an identical fashion (Table 4). These results are similar to the relative binding intensities measured by the quantitative ELISA assays.
| Antibody | Antigen | |||
|---|---|---|---|---|
| HSA | HL | TL | ||
| Polyclonal | Lane density | 1.05 × 108 | 1.45 × 108 | 1.99 × 107 |
| anti-Hevea | TKS Ratio | TL/HSA=0.19 | TL/HL=0.14 | |
| Monoclonal | Lane density | 9.37 × 106 | 2.94 × 106 | 2.79 × 105 |
| anti-Hev b 3 | TKS Ratio | TL/HSA=0.03 | TL/HL=0.09 | |
Table 4: Binding intensities of anti-Heveaantibodies toward Hevea and Taraxacum kok-saghyz latex proteins. Lane intensities as measured by chemiluminescent detection and image densitometry corresponding to Figures 1b and 1c are reported, followed by the calculated ratios of lane intensities for antibody recognition of TKS latex proteins relative to each Hevea latex protein sample.
Comparing TKS and other plant proteins for anti-Hevea polyclonal antibody cross-reactivity
The previous results suggested significantly reduced cross-reactivity of Hevea antibodies toward TKS latex proteins compared to Hevea latex proteins. However, decreased cross reactivity does not exclude the potential danger of TKS proteins in individuals previously sensitized to Hevea antigens. To evaluate the antigenicity of TKS latex proteins in relation to some commonly encountered plant-based foods, proteins from soybean (G. max), a non-rubber-producing plant, and the latex of romaine lettuce (L. sativa), a rubber-producing plant containing latex proteins that are homologous to Hevea latex proteins [26], but which has not been identified as a major cause of allergy (http://www.allergen. org/index.php), were selected to compare their relative extents of anti- Hevea antibody cross-reactivity.
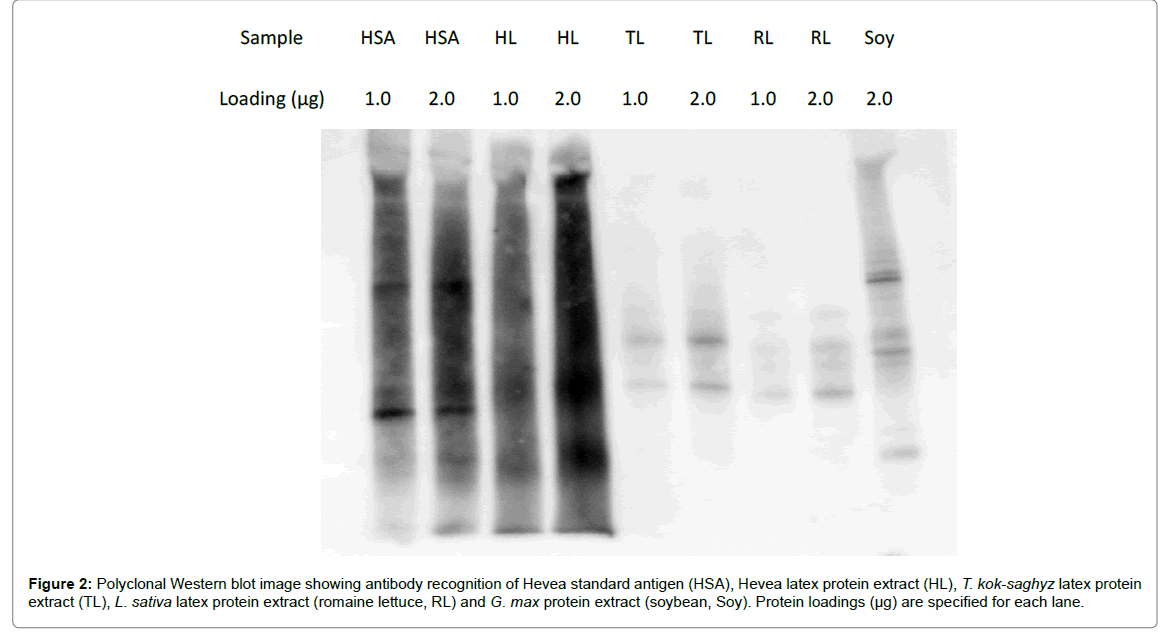
Figure 2: Polyclonal Western blot image showing antibody recognition of Hevea standard antigen (HSA), Hevea latex protein extract (HL), T. kok-saghyz latex protein extract (TL), L. sativa latex protein extract (romaine lettuce, RL) and G. max protein extract (soybean, Soy). Protein loadings (µg) are specified for each lane.
Qualitative observation of the Western blot image in Figure 2 showed that HSA and HL exhibited similarly strong reactive signals, while TKS latex proteins, lettuce latex proteins and soy proteins all showed significantly reduced signals. The lettuce latex proteins gave a similar binding intensity to that of TKS latex proteins and, interestingly, soy proteins produced a stronger signal than either the TKS or lettuce latex proteins.
When the relative intensities of antibody binding between other plant proteins and Hevea latex proteins were measured densitometrically, the intensity of TKS latex protein binding was 152% that of lettuce latex protein, 48% that of soy protein, but only 10% that of Hevea latex protein. Compared to Hevea latex protein, the relative intensities of lettuce and soy protein cross-reactivity were 7% and 21%, respectively (Table 5).
| Protein source (loading) | HSA (2.0 µg) | HL (2.0 µg) | TL (2.0 µg) | RL (2.0 µg) | Soy (2.0 µg) |
|---|---|---|---|---|---|
| Lane densities | 4.94 × 107 | 8.91 × 107 | 9.03 × 106 | 5.95 × 106 | 1.88 × 107 |
| Relative intensity ratios | |||||
| HSA | 0.55 | 0.18 | 0.12 | 0.38 | |
| HL | 1.81 | 0.1 | 0.07 | 0.21 | |
| TKS | 0.18 | 10 | 0.66 | 2.08 | |
| RL | 8.31 | 14.98 | 1.52 | 3.16 | |
| Soy | 2.63 | 4.75 | 0.48 | 0.32 |
Table 5: Binding intensities of anti-Hevea polyclonal antibody toward various plant proteins, relative to Hevea latex proteins. Values are expressed as the ratios of lane intensities from Figure 2 as measured by chemiluminescent detection.
These results demonstrate that while some cross-reactivity is apparent toward TKS latex proteins, the intensity of polyclonal anti- Hevea antibody binding toward TKS latex proteins is significantly lower than toward Hevea latex proteins and similar to or lower than some common plant foods including romaine lettuce and soybean.
Discussion
The rubber particle of H. brasiliensis contains at least 186 proteins based on proteomic analysis [27], while the whole cytoplasm latex of H. brasiliensis contains over 300 proteins [24], including 15 that have been identified thus far as being clinically significant in human type I latex allergy. These 15 proteins represent the canonical Hevea allergens, named Hev b 1 through Hev b 15 by the World Health Organization International Union of Immunological Studies (IUIS) (http://www. allergen.org/index.php). Hev b 1, Rubber elongation factor (HbREF, 14.7 kDa) and Hev b 3, Small rubber particle protein (HbSRPP, 22.3 kDa), are recognized as major allergens found in Hevea latex [28-32].
Phylogenetic analysis shows that HbREF and HbSRPP are homologous proteins originating from a common ancestor gene, and sequence analysis shows that they share highly conserved motifs with 50% sequence identity. Compared to HbSRPP, HbREF partially lacks C-terminal sequence [19]. This REF/SRPP family of proteins is widely distributed among members of the plant kingdom, denoted as SRPP or REF-like proteins in the UNIPROT database (http://www.uniprot.org/ uniprot/Q9MA63).
Cross-reactivity in Hevea-sensitive individuals toward various plant foods is due to the presence of homologous proteins [33]. Homologs of Hevea SRPP also exist in the rubber-producing plants Parthenium argentatum (guayule) (GHS) [34], T. kok-saghyz (TkSRPP) [35] and L. sativa (lettuce) (LsSRPP) [26]. To investigate the abundance of Hevea antigen homologs in both rubber-producing and non-rubberproducing plants, we performed BLAST queries (https://blast.ncbi.nlm. nih.gov/Blast.cgi) using the 15 Hevea allergens currently identified to search for homologous protein sequences from three other plant species: T. kok-saghyz, L. sativa (lettuce) and G. max (soybean). Redundant homologous protein sequences obtained from the search results were removed and the top five were tabulated for each plant species based on their identity scores, given in Supplementary Information.
The proteomic analysis identified different numbers of Hevea homologs among the different plants, with varying degrees of identity for each Hev antigen (Supplementary Information). As expected, due to their highly conserved sequence similarity, Hev b 1 and Hev b 3 share common homologous proteins of similar sequence identity with TKS. Three Hev b 3 homologs were identified in lettuce, two of which also share significant homology with Hev b 1. In soybean, at least five homologous proteins were identified for each of Hev b 1 and Hev b 3, of which two were shared. Homologs of Hev b 2, Hev b 4, Hev b 7, Hev b 8, Hev b 9, Hev b 13, and Hev b 15 were only identified in soybean. Hev b 5 gave no hits in any of the plants compared. Homologous proteins to Hev b 6, Hev b 10, Hev b 11, and Hev b 12 were only identified in lettuce and soybean. Hev b 14 had many high identity proteins shared with soybean and one low identity protein each in TKS and lettuce.
Complete genome sequences are available for lettuce and soybean, but not for TKS, which has only 263 protein sequences and 103 translated mRNA sequences available for comparison. It is interesting to note that lettuce contains proteins which are homologous to 7 out of the 15 Hevea antigens, but has no proteins which are homologous to the other 8 Hevea antigens. This may be why the weakest reactivity of the polyclonal anti-Hevea antibody was observed against lettuce proteins. Although only five proteins in TKS were identified as being homologous to the Hevea antigens, it is highly possible that additional homologous proteins will be identified in TKS when more genomic or proteomic data are available in the future.
Remarkably, a large number of homologs to 14 of the 15 Hevea antigens were identified in G. max, a non-rubber-producing plant. This homology is supported by the Western blot densitometric analysis which also showed the greatest amount of polyclonal antibody crossreactivity toward soybean proteins among the plants analyzed (Figure 2 and Table 5).
Soybean is ranked as one of the top 8 allergenic foods, especially among infants and children and at least 21 allergenic soybean proteins have been identified [36]. Approximately 0.4% of children are allergic to soy and among them the majority will outgrow the soy allergy by ages 3 to 10 [37]. Allergic reactions to soy are typically mild and only on rare occasions were severe reactions reported. Compared to many other food proteins, both human clinical and animal model data indicate that soy proteins tend to be less immunogenic: the minimum oral allergen dose required to initiate allergic symptoms (food allergen reaction thresholds) for soy is >100-fold greater relative to peanut, hazelnut, egg and milk. The safe protein dose for soy is approximately 400 mg whereas the other antigens range from 0.1 to 3 mg [38].
Our results suggest that TKS cross-reactivity may be analogous to the cross-reactivity to homologous proteins of some pollens and fruits in a portion of Hevea latex-allergic individuals. Approximately 30-50% of individuals allergic to Hevea latex show an associated hypersensitivity to some plant-derived foods, a condition called “latexfruit syndrome”. It has been hypothesized that allergen cross-reactivity is due the existence of structurally similar epitopes on different proteins that are phylogenetically closely related or represent evolutionarily conserved structures [33,39].
Therefore, while TKS latex proteins showed in vitro cross-reactivity by Hevea antibody recognition, the threat that TKS latex holds in vivo for latex allergic individuals may not be as severe as Hevea natural rubber latex; rather, it may fall within the range of cross-reactive food allergens reported to be associated (clinically or immunochemically) with natural rubber latex within the group of “low or undetermined” degree of association or prevalence by the American Latex Allergy Association (http://latexallergyresources.org/cross-reactive-food). This group contains soybean and many other fruits and vegetables, but as of yet does not include lettuce.
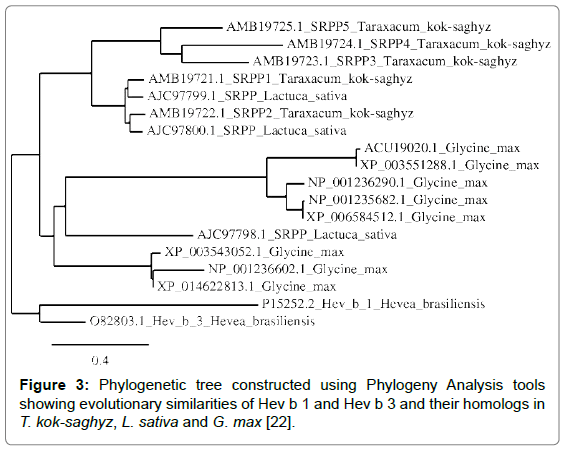
A phylogenetic tree was constructed to compare the evolutionary relationships of proteins identified as being homologous to Hev b 1 and Hev b 3, the Hevea allergens having the greatest number of homologs in the compared plant species (Figure 3).
Phylogenetic analysis shows that all homologs in G. max originate from a single ancestor, as does one lettuce SRPP protein, whereas all TKS latex proteins share a common ancestor with two other L. sativa latex proteins.
Based on the evolutionary distances of Hev b 1 and Hev b 3, and their homologs in other plants, G. max proteins are more closely related to L. sativa latex proteins than to H. brasiliensis latex proteins; similarly, TKS latex proteins are also more closely related to L. sativa latex proteins than to H. brasiliensis latex proteins, although this entire group of proteins likely originates from a common ancestor. Therefore, although TKS is a rubber-producing plant, in terms of potential allergenicity, TKS latex proteins may be more similar to proteins found in plant foods such as soy and lettuce, than to the Hevea allergens.
Latex-fruit syndrome is considered to be a class 2 food allergy caused by incomplete, labile food allergens that can only function as non-sensitizing elicitors; such food allergens elicit an immune response from antibodies developed against a complete antigen (class 1) having heat stability and resistance to digestive enzymes and which can function as both sensitizer and elicitor (http://dmd.nihs.go.jp/latex/ cross-e.html).
Our results cannot exclude the possibility that TKS proteins may contain some class 1 antigens which could induce sensitivity in nonallergic people, but the similarity of cross-reactivity and evolutionary relationships to other plant foods suggest that TKS latex may contain class 2 antigens, which cross-react mildly to Hevea antibodies and also may have much reduced potential to either induce de novo allergy or allergic reactions in Hevea allergic individuals.
As a rubber-producing plant having potential as a novel source of domestic natural rubber latex, TKS should be clinically evaluated not only for its cross-reactivity to H. brasilienis type I latex allergy, but also for its potential to induce type I allergies of its own [40]. Antigen exposure from latex products is mainly through skin contact or inhalation of airborne particles carrying latex antigens, which is distinct from oral and digestive exposure to food antigens. Further clarification of our in vitro results requires subsequent in vivo studies on TKS latex protein allergenicity using an animal model for human type I allergies.
Hypoallergenic natural rubber from sources other than Hevea has been reported in both non-laticiferous rubber-producing species such as P. argentatum (guayule) and laticiferous plants such as Ficus elastica (Cornish K, US Patent US5717050). As such, it has been suggested that Hevea latex allergens are species-specific, and Hevea latex allergy may be circumvented using rubber from divergent species [41].
Among three evolutionarily divergent rubber-producing plant species (Hevea, Taraxacum and Parthenium), latex-associated proteins originate from common ancestry within a larger family of plant stressrelated proteins (prenyl transferases) and share significant homology with a wide range of plant stress response proteins. It is expected that nonspecific antibody cross-reactivity occurs among these families of proteins [31]. However, direct comparison of the relative degree of antigenicity using quantitative methods provides an indication of the relative allergenicity of alternative natural rubber latex sources.
In conclusion, our results suggest that TKS may be an additional potential source for either hypoallergenic or circumallergenic natural rubber latex. These results do not indicate an absence of allergenicity upon exposure to TKS latex in Hevea-sensitized individuals, but emphasize the need for substantiation by in vivo testing.
Acknowledgement
The authors are grateful to Zerihun Demissie, Pierre Fobert, Susan Logan, and Jamshid Tanha for their constructive reviews of the manuscript and to Chelsea Barna for the summary diagram artwork.
Conflict of Interest
Funding for this work was provided by NovaBioRubber Green Technologies Inc. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare no competing financial interest.
References
- Gerber JF (2011) Conflicts over industrial tree plantations in the South: Who, how and why? Glob Environ Change 1: 165-176.
- de Blécourt M, Brumme R, Xu J, Corre MD, Veldkamp E (2013) Soil carbon stocks decrease following conversion of secondary forests to rubber (Hevea brasiliensis) plantations. PLoS ONE 8: e69357.
- Farrell BD, Dussourd DE, Mitter C (1991) Escalation of plant defense: Do latex and resin canals spur plant diversification? Am Nat 4: 881-900.
- Konno K (2011) Plant latex and other exudates as plant defense systems: Roles of various defense chemicals and proteins contained therein. Phytochemistry 13: 1510-1530.
- Cornish K, Siler DJ (1996) Hypoallergenic guayule latex: Research to commercialization. Prog New Crops ASHS Press Alex VA: 327–335.
- Palosuo T, Antoniadou I, Gottrup F, Phillips P (2011) Latex medical gloves: Time for a reappraisal. Int Arch Allergy Immunol 156: 234-246.
- Nanti S, Wongputtisin P, Sakulsingharoj C, Klongklaew A, Chomsri N (2014) Removal of allergenic protein in natural rubber latex using protease from Bacillus sp. Food Appl Biosci J 3: 216-223.
- Perera ALHA, Perera BGK1 (2017) Development of an economical method to reduce the extractable latex protein levels in finished dipped rubber products. Biomed Res Int 2017: 9573021.
- Palosuo T1, Alenius H, Turjanmaa K (2002) Quantitation of latex allergens. Methods 27: 52-58.
- Palosuo T, Reinikka-Railo H, Kautiainen H, Alenius H, Kalkkinen N, et al. (2007) Latex allergy: The sum quantity of four major allergens shows the allergenic potential of medical gloves. Allergy 7: 781–786.
- Buranov AU, Elmuradov BJ (2010) Extraction and characterization of latex and natural rubber from rubber-bearing plants. J Agric Food Chem 58: 734-743.
- Cornish K, Xie W (2015) Immunological analysis of the alternate rubber crop Taraxacum koksaghyz indicates multiple proteins cross-reactive with Hevea brasiliensis latex allergens. J Biotechnol Biomater 4.
- Cornish K, Chapman MH, Nakayama FS, Vinyard SH, Whitehand LC (1999) Latex quantification in guayule shrub and homogenate. Ind Crops Prod 2: 121-136.
- Cornish K, Xie W (2011) Natural rubber biosynthesis in plants: Rubber transferase. Methods Enzymol 515: 63-82.
- Siler DJ, Cornish K (1995) Measurement of protein in natural rubber latex. Anal Biochem 229: 278-281.
- ASTM (2015) Standard test method for analysis of aqueous extractable protein in latex, natural rubber and elastomeric products using the modified Lowry method, D5712-15.
- ASTM (2012) Standard test method for the immunological measurement of antigenic protein in natural rubber and its products, D6499.
- ASTM (2014) Standard test method for immunological measurement of four principal allergenic proteins (hev b 1, 3, 5 and 6.02) in natural rubber and its products derived from latex, D7427-14.
- Berthelot K, Lecomte S, Estevez Y, Peruch F (2014) Hevea brasiliensis REF (Hev b 1) and SRPP (Hev b 3): An overview on rubber particle proteins. Biochimie 106: 1-9.
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680-685.
- Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci 9: 4350-4354.
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, et al. (2008) Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 2: W465-W469.
- Dereeper A, Audic S, Claverie JM, Blanc G (2010) BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol 10: 8.
- D’Amato A, Bachi A, Fasoli E, Boschetti E, Peltre G, et al. (2010) In-depth exploration of Hevea brasiliensis latex proteome and “hidden allergens” via combinatorial peptide ligand libraries. J Proteomics 7: 1368-1380.
- Beezhold DH, Kostyal DA, Tomazic-Jezic VJ (2002) Measurement of latex proteins and assessment of latex protein exposure. Methods 1: 46-51.
- Chakrabarty R, Qu Y, Ro D-K (2015) Silencing the lettuce homologs of small rubber particle protein does not influence natural rubber biosynthesis in lettuce (Lactuca sativa). Phytochemistry 113: 121-129.
- Dai L, Kang G, Li Y, Nie Z, Duan C, et al. (2013) In-depth proteome analysis of the rubber particle of Hevea brasiliensis (para rubber tree). Plant Mol Biol 1-2: 155-168.
- Yeang HY, Cheong KF, Sunderasan E, Hamzah S, Chewa NP, et al. (1996) The 14.6 kd rubber elongation factor (Hev b 1) and 24 kd (Hev b 3) rubber particle proteins are recognized by IgE from patients with spina bifida and latex allergy. J Allergy Clin Immunol 3: 628-639.
- Yeang H, Ward M, Zamri A, Dennis M, Light D (1998) Amino acid sequence similarity of Hev b 3 to two previously reported 27 and 23 kDa latex proteins allergenic to spina bifida patients. Allergy 5: 513-517.
- Yeang HY1, Arif SA, Yusof F, Sunderasan E (2002) Allergenic proteins of natural rubber latex. Methods 27: 32-45.
- Czuppon AB, Chen Z, Rennert S, Engelke T, Meyer HE, et al. (1993) The rubber elongation factor of rubber trees (Hevea brasiliensis) is the major allergen in latex. J Allergy Clin Immunol 5: 690-697.
- Berthelot K, Lecomte S, Estevez Y, Coulary-Salin B, Peruch F (2014) Homologous Hevea brasiliensis REF (Hevb1) and SRPP (Hevb3) present different auto-assembling. Biochim Biophys Acta BBA - Proteins Proteomics 2: 473-485.
- Wagner S, Breiteneder H (2002) The latex-fruit syndrome. Portland Press Limited.
- Kim IJ, Ryu SB, Kwak YS, Kang H (2004) A novel cDNA from Parthenium argentatum Gray enhances the rubber biosynthetic activity in vitro. J Exp Bot 55: 377-385.
- Collins-Silva J, Nural AT, Skaggs A, Scott D, Hathwaik U, et al. (2012) Altered levels of the Taraxacum kok-saghyz (Russian dandelion) small rubber particle protein, TkSRPP3, results in qualitative and quantitative changes in rubber metabolism. Phytochemistry: 46-56.
- Wilson S, Blaschek K, de Mejia E (2005) Allergenic proteins in soybean: Processing and reduction of P34 allergenicity. Nutr Rev 63: 47-58.
- Savage JH, Kaeding AJ, Matsui EC, Wood RA (2010) The natural history of soy allergy. J Allergy Clin Immunol 125: 683-686.
- Cordle CT (2004) Soy protein allergy: Incidence and relative severity. J Nutr 134: 1213S-1219S.
- Brehler R, Theissen U, Mohr C, Luger T (1997) Latex-fruit syndrome: Frequency of cross-reacting IgE antibodies. Allergy 4: 404-410.
- Cornish K, McMahan CM, Pearson CH, Ray DT, Shintani DK (2005) Biotechnological development of domestic rubber producing crops. Rubber World 2: 40-44.
- Siler DJ, Cornish K (1994) Hypoallergenicity of guayule rubber particle proteins compared to Hevea latex proteins. Ind Crops Prod 4: 307-313.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 3836
- [From(publication date):
September-2017 - Jul 06, 2025] - Breakdown by view type
- HTML page views : 2946
- PDF downloads : 890