Research Article Open Access
Proteome Analysis on Proteins Sequentially Extracted from Loach (Misgurnus anguillicaudatus)
Yanlei Yu1, Ning Zang2, Fuming Zhang3*, Robert J Linhardt3 and Hong Zhang1*
1School of Food Science and Biological Engineering, Zhejiang Gongshang University, China
2Medical Scientific Research Center, Guangxi Medical University, Nanning, China
3Departments of Chemistry and Chemical Biology, Center for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, New York, USA
- *Corresponding Author:
- Hong Zhang
School of Food Science and Biological Engineering
Zhejiang Gongshang University, Hangzhou
Zhejiang, 310018, China
Tel: +86-571-2800-8966
Email: hongzh1316@mail.zjgsu.edu.cn
Fuming Zhang
Department of Chemical and Biological Engineering
Center for Biotechnology and Interdisciplinary Studies
Rensselaer Polytechnic Institute, Troy
NY12180, USA.
Tel. +1-518-276-6839
E-mail: zhangf2@rpi.edu
Received date: February 04, 2017; Accepted date: March 27, 2017; Published date: April 03, 2017
Citation: Yu Y, Zang N, Zhang F, Linhardt RJ, Zhang H (2017) Proteome Analysis on Proteins Sequentially Extracted from Loach (Misgurnus anguillicaudatus). Biochem Physiol 6: 214. doi: 10.4172/2168-9652.1000214
Copyright: © 2017 Yu Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Visit for more related articles at Biochemistry & Physiology: Open Access
Abstract
Loach (Misgurnus anguillicaudatus) is known as a selenium-rich aquatic product. In present study, a sequential extraction method was applied for separation of water, salt and alkali-soluble proteins from loach (Misgurnus anguillicaudatus). The extracted proteins (included 12 water-soluble, 4 salt-soluble and 7 alkali-soluble proteins) were subjected to molecular weight analysis by SDS-PAGE and MALDI-TOF MS. Then, water soluble proteins were chosen for further ion exchange purification after selenium tracking by ICP-MS. The selenium containing peptides in water soluble proteins were identified by ESI-MS. Four proteins including creatine kinase, cytochrome P450 aromatase, beta-actin and glyceraldehyde 3-phosphate dehydrogenase were identified. Among these proteins, two selenium-containing peptides from beta-actin and cytochrome P450 aromatase were identified. The results of proteome analysis provide valuable molecular information of loach proteins. Moreover, it helps to understand the effect of selenium on redox systems in loach and its antioxidant functions.
Keywords
Misgurnus anguillicaudatus; Protein identification; Selenium; Sequential extraction
Abbreviations
EMS: Enhanced MS; EPI: Enhanced Product Ion; ER: Enhanced Resolution; GAPDH: Glyceraldehydes-3-Phosphate Dehydrogenase; GPx: Glutathione Peroxidase; ICP-MS: Inductively Coupled with Plasma-Mass Spectrometry; NCBI: National Center for Biotechnology Information; RDA: Recommended Dietary Allowance; Se-Met: Selenomethione; Se-Cys: Selenocysteine; Se-P: Selenoprotein P
Introduction
Selenium (Se) is an essential trace element that plays important roles of vital processes in human body. The recommended dietary allowance (RDA) of selenium for adults is 55 μg/day with a tolerable upper intake level of 400 μg/day [1]. The narrow concentration range between essentiality and toxicity requires a thorough understanding of selenium. Se is an important part of the antioxidant enzymes that protects cells against free radicals. It is also an essential component of major metabolic pathways, including the antioxidant defense systems, the immune system and thyroid hormone metabolism [2]. An adequate intake of selenium can decrease the incidence of diseases like heart disease, cancer, cardiovascular sclerosis and diabetes [3-7]. Selenium exists predominantly in plants and aquatic products in both inorganic forms, such as selenite, selenate, selenide and in organic forms like selenomethionine and selenocysteine. Aquatic products are the richest source of selenium and possess high nutritional value [8]. They have been considered an important source for selenium supplementation for decades [9-11].
Loach (Misgurnus anguillicaudatus) is a bottom-feeding freshwater fish in the superfamily Cobitoidea and is widely consumed in East Asian countries including China, Korea and Japan [12]. It was reported in the Dictionary of Chinese Traditional Medicine as a treatment for hepatitis, osteomyelitis, carbuncles, inflammations and cancers [13]. In our previous research, loach was determined to be a selenium-rich aquatic product. The selenium concentration in loach muscle is 0.444 ± 0.003 mg/kg, which is much higher than many other aquatic products [14]. Furthermore, loach possesses high nutritional and medicinal values and is an appropriate supplement for dairy products.
Protein-bound selenium has generated particular interest due to the important role Se plays in redox systems. Selenium is incorporated into proteins mainly in the form of selenomethionine (Se-Met) and selenocysteine (Se-Cys) as a part of antioxidant enzymes such as glutathione peroxidase (GPx) and selenoprotein-P (Se-P) [15]. Selenium is important for antioxidant defense as an essential component of these enzymes, and supplementation directly affects its activity. In our previous studies, selenium concentration and the content of Se-Met and Se-Cys in loach muscle were determined by HPLC-ICP-MS and HPLC-ESI-MS/MS. The results demonstrated that Se-Met was the predominant selenoamino acid. However, the location of selenium within proteins needs further analysis. In other research, You and coworkers investigated the antioxidant activity and antifatigue effects of loach protein hydrolysates [16,17]. They focused on amino acid analysis and antioxidant peptide identification providing an insight of loach protein profile and the connection between selenium and loach proteins. In the present study, a sequential extraction method was used to recover loach proteins. The protein profile and selenium concentration in water, aqueous sodium chloride, ethanol and sodium hydroxide fractions were determined. The water soluble protein was chosen for ion-exchange separation and further analysis of selenium distribution. Selenium containing peptides and proteins in water soluble proteins were identified, providing a better understanding of how selenium functions in redox system of loach.
Materials and Methods
Materials and chemicals
Live loach (Misgurnus anguillicaudatus) was purchased from a local market (Cuiyuan, Hangzhou, China). Fresh edible muscle tissues were collected and ground to homogeneity for further analysis. Formic acid and acetonitrile were of HPLC grade obtained from Merck (Germany), DEAE-Sephacel was obtained from Amersham Bio-Sciences (USA), Trypsin was obtained from Sigma (USA). All other chemicals used were of analytical grade.
Protein extraction
Loach muscle tissues were sequentially extracted with water, aqueous sodium chloride, ethanol and sodium hydroxide. Briefly, tissues were defatted twice using -40ºC precooled acetone, then the 20 g defatted samples were mixed with deionized water at 4ºC in a ratio of 1:20 (w/v) and stirred for 4 h. The supernatant was collected after centrifugation at 10,000 g at 4ºC for 15 min and -40ºC precooled acetone was added slowly to get a final concentration as 80% saturation. The mixture was kept at 4ºC overnight. The precipitate was collected after centrifugation at 8,500 g at 4ºC for 15 min, re-dissolved and dialyzed against deionized water and then lyophilized for further purification. The procedure for the extraction of the other three protein fractions used the same approach except in place of water relied on: 0.5 mol/L sodium chloride for salt-soluble proteins, 75% (v/v) ethanol/water for alcohol-soluble proteins, and 0.25 mol/L sodium hydroxide for alkali-soluble proteins.
Selenium concentration determination
Selenium concentration was determined by ICP-MS (X-series II; Thermo Fisher, USA). Samples (0.25 g for fresh and 0.10 g for dried loach tissues) were digested in a mixture of 2 mL hydrogen peroxide and 4 mL nitric acid solution using a HP 500 vessel in Microwave Digestion system (MARS; CEM, USA). The digestion mixture was heated to 120ºC in 5 min and maintained for 2 min, then heated to 180ºC in 5 min and maintained for 15 min. Supernatants from the digestion were analyzed by ICP-MS under optimum conditions using 72Ge and 89Y as double internal standards.
Ion-exchange chromatography of water-soluble protein
The dried water-soluble protein fraction (100 mg) was re-dissolved in 20 mmol/L Tris-HCl buffer (pH 8.0) a final concentration of 20 mg/ mL and loaded onto DEAE-Sephacel anion-exchange column, which was previously equilibrated with the same buffer overnight. The column was eluted at a flow rate of 0.6 mL/min, with a stepwise gradient elution of water and 1.5 column volumes each of 20 mmol/L Tris-HCl buffer containing 0.1, 0.3, 0.5 and 1 mol/L sodium chloride. Every 6 mL of eluted solution was collected and absorbance was monitored at 280 nm. The fractions corresponding to the desire peaks were pooled, dialyzed (3.5 kDa), concentrated, and lyophilized for further use. The selenium concentration of each fraction was determined by ICP-MS.
SDS-PAGE analysis and in-gel digestion
Protein samples were mixed with an equal volume of double strength sample loading buffer, and boiled for 3 min. Electrophoresis was carried out at room temperature, 80 V in voltage of stacking gel, 120 V in voltage of running gel, and the gel was stained with Coomassie Brilliant Blue R250 for 20 min, followed by Destain in destaining solution containing 10% methanol and 10% acetic acid (v/v). In-gel digestion was performed by the protocol of Shevchenko [18]. The main procedure included excising protein bands from stained gels and cutting these into small pieces about 1 mm3, transferring to a 0.8 mL Eppendorf centrifuge tube, and in-gel reduction and alkylation, digested with trypsin overnight, finally the desalted peptide was stored for MS analysis.
Mass spectrometry analysis
Four soluble proteins were analyzed by MALDI-TOF/TOF MS. Acquisition parameters, resolution and laser energy was optimized prior to MS analysis. The instrument was operated in positive ionization mode. MS conditions of 1000 shots per spectrum were used and MS/MS was performed with 4000 laser shots per spectrum. Watersoluble protein was analyzed by HPLC-ESI-MS/MS. Chromatographic separations were performed on a LC sync system with aqueous buffer A (0.1% formic acid) and buffer B (90% acetonitrile in 0.1% formic acid) with gradient elution in 40 min. Mass spectra were recorded in positive ionization mode. Data were collected in profile mode using one Enhanced MS (EMS) scan, followed by an Enhanced Resolution (ER) scan and three Enhanced Product Ion (EPI) scans.
Data analysis
Protein identification was conducted against a database search in non-redundant National Center for Biotechnology Information (NCBI) using MASCOT.
Results
Protein distribution and selenium concentration after sequential extraction
Protein extraction relied on a sequential extraction method. Loach muscle tissues were sequentially extracted by water, aqueous sodium chloride, ethanol and sodium hydroxide solutions. The protein fractions obtained and their selenium concentrations are shown in Table 1. The alkali-soluble protein fraction was the most abundant and water-soluble protein fraction was the most selenium-rich. No ethanolsoluble protein was obtained.
SDS-PAGE and protein identification by MALDI TOF MS
The SDS-PAGE analysis results (Figure 1) showed the bands and their molecular weights of proteins present in the water, alkali, and salt soluble fractions. Based on the bands observed, 7 bands from watersoluble protein, 6 bands from salt-soluble protein, and 6 bands from alkali-soluble protein were recovered for in-gel digestion and mass spectrometric analysis. In the water soluble protein fraction, there were 12 proteins identified that matched the molecular weight results obtained by SDS-PAGE. Three proteins around 42 kDa and three proteins around 35 kDa were identified. In the salt soluble protein fraction, only 4 proteins were identified that matched the results obtained by SDSPAGE. In the alkali-soluble protein fraction, the separation was poor and the background was complicated, but 7 proteins were matched to the SDS-PAGE analysis. The matched proteins of water, alkali and saltsoluble protein fractions are shown in Table 2 and Figure 1.
| Loach muscle tissues (15 g) | ||||
|---|---|---|---|---|
| Water-soluble protein | Salt-soluble protein | Ethanol-soluble protein | Alkali-soluble protein | |
| Extraction times | 3 | 2 | 1 | 4 |
| mass (mg) | 491.00 | 247.03 | 0 | 1271.11 |
| Selenium concentration (mg/kg, n=3) | 0.165 ± 0.001 | 0.027 ± 0.006 | 0 | 0.061 ± 0.009 |
Table 1: Protein distributions and its selenium concentrations.
| Number | Accession | Description | Mass (Da) | Score | Peptide no. |
|---|---|---|---|---|---|
| Water-soluble proteins | |||||
| 1 | gi|4027927 | Creatine kinase M2-CK [Cyprinus carpio] | 42900.6 | 460 | 16 |
| 2 | gi|307548819 | Cytochrome P450 aromatase [Misgurnus anguillicaudatus] |
45206.2 | 123 | 12 |
| 3 | gi|119943232 | Beta-actin [Misgurnus anguillicaudatus] | 42369.6 | 109 | 11 |
| 4 | gi|658865028 | Keratin, type I cytoskeletal 13-like isoform X2 [Poecilia reticulate] | 54984 | 99 | 28 |
| 5 | gi|119943230 | Glyceraldehydes 3-phosphate dehydrogenase [Misgurnus anguillicaudatus] | 35894.8 | 89 | 9 |
| 6 | gi|410922980 | Trichoplein keratin filament-binding protein-like [Takifugu rubripes] | 61762 | 74 | 19 |
| 7 | gi|55669145 | Lactate dehydrogenase B-type subunit [Cyprinus carpio] |
36498 | 71 | 16 |
| 8 | gi|551516116 | Zinc finger protein 316-like [Xiphophorus maculates] |
38356 | 68 | 19 |
| 9 | gi|308321302 | Parvalbumin beta [Ictalurus furcatus] | 11625 | 68 | 7 |
| 10 | gi|528491535 | Heme oxygenase-like [Oryzias latipes] | 31042 | 66 | 17 |
| 11 | gi|657757727 | FGFR1 oncogene partner 2 [Cynoglossus semilaevis] | 25687 | 63 | 12 |
| 12 | gi|528516409 | Serine/threonine-protein kinase pim-2-like [Danio rerio] | 36382 | 52 | 12 |
| Salt-soluble proteins | |||||
| 1 | gi|658853627 | Cellular retinoic acid-binding protein 1 [Poecilia reticulate] | 15524 | 71 | 12 |
| 2 | gi|584029529 | Protein asteroid homolog 1-like [Neolamprologus brichardi] | 58224 | 66 | 15 |
| 3 | gi|542250972 | SKI family transcriptional corepressor 1 homolog-B-like isofoerm X2 [oreochromis niloticus] |
85368 | 64 | 10 |
| 4 | gi|499047087 | 7SK snRNA methylphosphate capping enzyme-like [Maylandia zebra] | 69044 | 62 | 15 |
| Alkali-soluble proteins | |||||
| 1 | gi|548352670 | GTPase IMAP family member 7-like [Pundamilia nyererei] | 34822 | 58 | 7 |
| 2 | gi|658884301 | Eukaryotic translation initiation factor 2A [Poecilia reticulata] | 63920 | 65 | 12 |
| 3 | gi|584005499 | Peroxisomal NADH pyrophosphatase NUDT12-like [Neolamprologus brichardi] | 50847 | 53 | 11 |
| 4 | gi|657534298 | Tropomyosin alpha-4 chain isoform X1 [Stegastes partitus] | 32679 | 52 | 8 |
| 5 | gi|410922980 | Trichoplein keratin filament-binding protein-like [Takifugu rubripes] | 61762 | 69 | 18 |
| 6 | gi|657791047 | Dihydropyrimidinase [Cynoglossus semilaevis] | 54228 | 59 | 11 |
| 7 | gi|410917574 | Gem-associated protein 6-like [Takifugu rubripes] |
18198 | 61 | 10 |
Table 2: Identified loach proteins by MALDI TOF MS.
Figure 1: SDS-PAGE analysis and identified proteins from three soluble protein extractions.
Lane A: MW marker
Lane B: Water-soluble protein, proteins identified from bands (indicated with arrows), B-1: Keratin; B-2: Cytochrome P450 aromatase, creatine kinase M2-CK and betaactin;
B-3: Glyceraldehydes 3-phosphate dehydrogenase, lactate dehydrogenase and zinc finger protein; B-4: FGFR1 oncogene; B-5: Parvalbumin
Lane C: Salt-soluble protein, proteins identified from bands (indicated with arrows), C-1: SKI family transcriptional corepressor; C-2: 7SK snRNA methylphosphate capping enzyme; C-3: Protein asteroid homolog; C-4: Cellular retinoic acid-binding protein
Lane D: Alkali-soluble protein, proteins identified from bands (indicated with arrows), D-1: Eukaryotic translation initiation factor; and trichoplein keratin filament-binding
protein; D-2:Dihydropyrimidinase and peroxisomal NADH pyrophosphatase; D-3: GTPase IMAP family member and tropomyosin alpha-4 chain Figure 1: SDS-PAGE analysis and identified proteins from three soluble protein extractions.
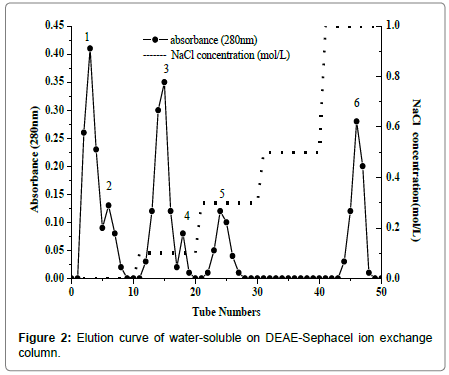
Purification of water soluble proteins and Se concentration in each fraction
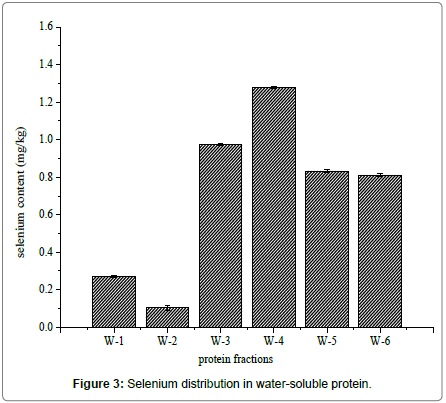
Water-soluble proteins were further purified by Ion exchange chromatography to determine the relationship between selenium and antioxidant proteins. There were six major fractions in water-soluble protein after DEAE Sephacel ion exchange chromatography separation (Figure 2). The six peaks were labeled W-1 to W-6. The selenium distribution in the purified water soluble proteins fractions is presented in Figure 3. Selenium concentration reached was the highest, 1.278 ± 0.005 mg/kg, in W-4, which eluted at 0.1 mol/L sodium chloride. The selenium content was lowest, 0.270 ± 0.004 mg/kg and 0.103 ± 0.011 mg/kg, in W-1 and W-2, respectively. W-3 had the second highest selenium content, 0.973 ± 0.006 mg/kg, and W-5 and W-6 followed closely at 0.761 ± 0.006 mg/kg and 0.844 ± 0.009 mg/kg, respectively.
Selenium-containing peptides identification by ESI MS
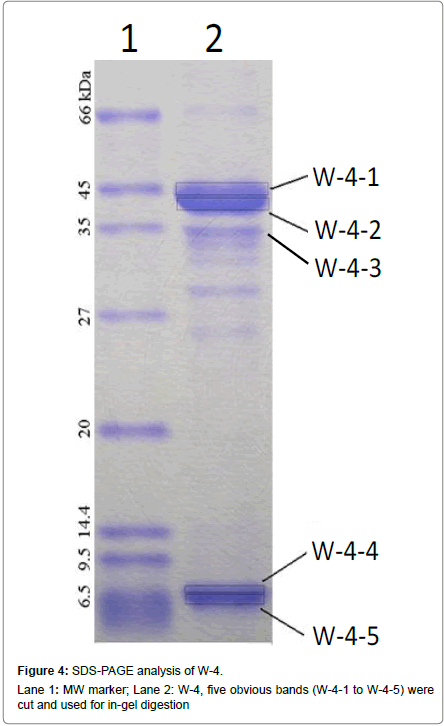
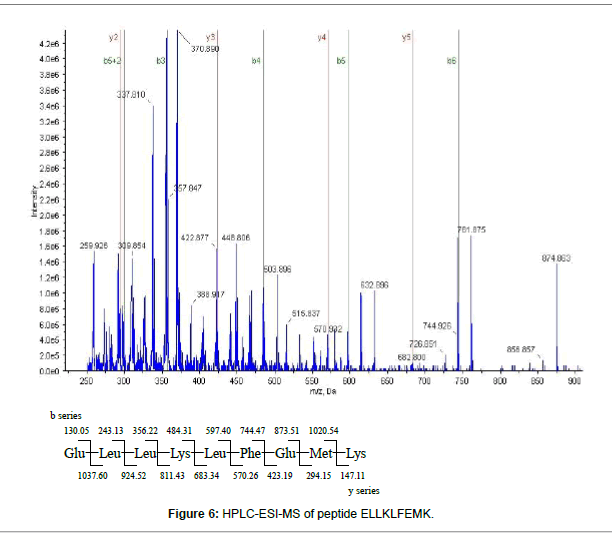
The W-4 fraction was subject to SDS-PAGE analysis (Figure 4). The 5 bands were cut and run in-gel digested and analyzed by HPLC-ESIMS. Four proteins including: Creatine kinase (43 kDa), Cytochrome P450 aromatase and Beta-actin (42-43 kDa) were found in W-4- 1bands, Glyceraldehyde 3-phosphate dehydrogenase (37 kDa) was found in W-4-2 band. No protein was identified in W-4-3, W-4-4 and W-4-5. Among the 4 identified proteins, two selenium-containing peptides from cytochrome P450 aromatase and beta-actin were identified from a manual search of the database. The mass spectrum of the peptide EITSLAPSTMK, is shown in Figure 5A. The Y3 ion (m/z 379.20, TMK) was present at m/z 378.9014 in Figure 5B. A selenium analogue at M+48/z and its isotopic pattern, corresponding to 78Se and 82Se, provide validation based on the selenium isotope pattern, m/z 425.1100, 426.8903 and 428.0209, as shown in the inset. The fragment of Y4 ion (m/z 466.23, STMK) is shown in Figure 5C with m/z 465.8938, its selenium substitute and its isotopes at m/z 511.7456 and 513.3600. In Figure 5D, the Y5 ion (m/z 563.29, PSTMK) and its M+48/z are observed at m/z 611.0918. In Figure 5E, the 6Y ion (m/z 634.32, APSTMK), 6Y+48/z (m/z 683) and its isotope (m/z 681.9986, 683.8201) are observed. Another peptide ELLKLFEMK (Figure 6) from cytochrome P450 aromatase was identified using the same methods.
Discussion
A sequential extraction method was applied for loach protein isolation. The three proteins obtained were separated by SDS-PAGE and identified by MALDI-TOF MS. The selenium content was determined and identify their selenium containing peptide sequences. Water soluble protein fraction was further purified by ion exchange chromatography and analyzed by ESI-MS. SDS-PAGE represented the ideal way to separate these selenoproteins [19]. While SDS-PAGE affords high resolution, there might be several proteins within a single band. This is a particular concern for the water-soluble protein fractions where, there were three proteins Creatine kinase, Cytochrome P450 aromatase and Beta-actin at around 42 kDa and Glyceraldehyde 3-phosphate dehydrogenase, Lactate dehydrogenase and Zinc finger protein at around 35 kDa.
Among the proteins identified by MALDI TOF MS, we focused on Beta-actin and Creatine kinase. Beta-actin is a highly conserved eukaryotic protein that exists in different cellular isoforms and its expression can change in response to biochemical stimuli during growth and differentiation and in disease states that may play a prominent role in cancer. Beta-actin constitutes a direct target for oxidative modification in pathophysiological states [20]. Glutathionylation is an important redox dependent mechanism for the regulation in Beta-actin [21]. In our previous studies, we found that loach has higher selenium content than other aquatic products. Selenium is an important micronutrient in redox center and has significant functions best-known for antioxidant activities as a constituent of selenoproteins [22]. Beta-actin may be connected to selenium concentration. Creatine kinase is an enzyme expressed by various tissues and cell types, it catalyses the conversion of creatine and adenosine triphosphate to creatine phosphocreatine and adenosine diphosphate. Arthur found that selenium status affects plasma creatine kinase activity in calves [23]. Keratin is the key structural material making up the outer layer of human skin. Sulfur supplements cause skin, hair and nails to grow faster, sulfur and selenium are antagonists, so selenium may also impact keratin. Lactate dehydrogenase is an enzyme that catalyzes the conversion of pyruvate to lactate. Kaur found that a selenium deficient diet may be responsible for reduced levels of lactate dehydrogenase [24]. Heme oxygenase is an enzyme that catalyzes the degradation of heme. Selenium is a novel regulator of cellular heme metabolism inducing the microsomal enzyme heme oxygenase [25]. The other proteins identified in this study were not known to be involved with selenium metabolism.
The novel findings of the study include manual searching based on selenium isotope distributions and the molecular weight differences between selenium and sulfur (m/z 48). Before analysis is performed, it is necessary to properly prepare samples in high purity without changing the chemical forms of the element [26]. In the present study, we suitably chose ion exchange chromatography and used ICP-MS method for monitoring selenium distribution. Another advantage of tracking selenium was choosing selenium-rich protein fractions for further identification. As selenium is covalently bound, enzymatic hydrolysis has provided a dramatic reduction of selenium-containing peptides with negligible degradation [27]. Two selenium peptides were found in cytochrome P450 aromatase and beta-actin protein, respectively. The mass spectra allow the identification of a seleniumcontaining peptide in each fraction due to selenium substitution and its isotopes. Selenomethionine (Se-Met) can randomly incorporate into proteins and substitute for methionine (Met) through a non-specific pathway [28,29]. Thus, the fractions of sulfur-peptide and a subset of their selenium analogs co-exist in peptides and their molecular weight differ by m/z 48 (32S and 80Se). These are observed in B-ions and Y-ions analyzed by nano-HPLC-ESI-MS/MS [30,31]. Selenium also has a unique isotope distribution of 74Se (0.89%), 76Se (9.36%), 77Se (7.63%), 78Se (23.78%), 80Se (49.61%) and 82Se (8.73%) in nature and this provides a “finger print” of the presence of Se in peptide [32-34], especially the presence of a signal between the peaks corresponding to 78Se, 80Se and 82Se [35].
As a result, selenium-containing peptides EITSLAPSTMK from beta-actin and ELLKLFEMK from Cytochrome P450 arornatase were identified. The results provided novel insights into the effect of selenium in redox system of loach protein. For peptide from beta-actin, experiments have indicated that beta-actin could constitute a direct target for oxidative modification in pathophysiological states [20]. As to selenium dependent enzymes such as glutathione peroxidases, thioredoxin reductases, and iodothyronine deiodinases can protect organisms against oxidation damage from free radicals and reactive oxygen species. Evidence also showed that selenium was involved in the GSH-dependent metabolism of hydroperoxides [36] and that glutathionylation is an important redox dependent mechanism of regulation in beta-actin. A possible explanation for the molecular basis of redox-regulated in beta-actin could be that sulfur had been substituted by selenium in peptides. Thus, this protein showed significant antioxidant activities. As for the other peptide identified, the mechanism of bound selenium remains unclear and requires further studies.
Conclusion
In conclusion, a sequential extraction method was applied for loach protein isolation. There were 12 water-soluble, 4 salt-soluble and 7 alkali-soluble proteins identified by MALDI-TOF MS that matched molecular weight analysis by SDS-PAGE. Water soluble protein was chosen for further purification after selenium tracking by ICP-MS. A novel insight into the effect of selenium in antioxidant of water soluble protein was provided and two selenium-containing peptides, EITSLAPSTMK from beta-actin and ELLKLFEMK from cytochrome P450 aromatase were identified. The results of proteomics provide a valuable resource for molecular analysis of loach proteins and help us better understand the affect of selenium on redox systems in loach and its antioxidant functions.
Acknowledgement
This work was supported by International Science and Technology Cooperation Program of China (2012DFA31250-1).
References
- Santhosh KB, Priyadarsini KI (2014) Selenium nutrition: How important is it? Biomedicine and Preventive Nutrition 4: 333-341
- Brown KM, Authur JR (2001) Selenium, selenoproteins and human health: A review. Public Health Nutr 4: 593-599.
- Salvini S, Diet D, Hennekens C (1995) Plasma levels of the antioxidant selenium and risk of myocardial infarction among U.S. physicians. Am J Cardiol 76: 1218-1221
- Tanguy S, Grauzam S, Leiris JD, Boucher F (2012) Impact of dietary selenium intake on cardiac health: Experimental approaches and human studies. Mol Nutr Food Res 56: 1106-1121
- Tinggi U (2008) Selenium: Its role as antioxidant in human health. Environ Health Prev Med. 13: 102-108
- Soriano-Garcia M (2004) Organoselenium compounds as potential therapeutic and chemopreventive agents: A review. Curr Med Chem 11: 1657-1669
- Li T, Yang W, Li M, Byun DS, Tong C, et al. (2008) Expression of selenium-binding protein 1 characterizes intestinal cell maturation and predicts survival for patients with colorectal cancer. Mol Nutr Food Res 52: 1289-1299
- Rayman MP (2008) Food-chain selenium and human health: Emphasis on intake. Brit J Nutr 100: 254-268
- Kieliszek M, Blazejak S (2013) Selenium: Significance, and outlook for supplementation. Nutr 29: 713-718.
- Pedrero Z, Madrid Y (2009) Novel approaches for selenium speciation in foodstuffs and biological species: A review. Anal Chim Acta 634: 135-152
- Yu Y, Zhang F, Lu D, Zhang H (2014) Selenium bioavailability from shrimps (Penaeus vannamei Boone) and its effect on the metabolism of phospholipid and cholesterol ester. J Funct Foods 6: 186-195
- Franch N, Clavero M, Garrido M, Gaya N, Lopez V, et al. (2008) On the establishment and range expansion of oriental weather fish (Misgurnus anguillicaudatus) in NE Iberian Peninsula. Biol Invasions 10: 1327-1331
- Zhang CX, Huang KX (2006) Mechanism of apoptosis induced by a polysaccharide, from the loach Misgurnus anguillicaudatus (MAP) in human hepatocellular carcinoma cells. Toxicol Appl Pharm 210: 236-245.
- Gong L, Xu Q, Lee C, Zhang H (2012) Selenium speciation analysis of Misgurnus anguillicaudatus selenoprotein by HPLC-ICP-MS and HPLC-ESI-MS/MS. Eur Food Res Technol 235: 169-176
- Rotruck JT, Pope AL, Ganther HE, Swanson AB, Hafeman DG, et al. (1973) Selenium: Biochemical role as a component of glutathione peroxidase. Science 179: 588-590
- You L, Zhao M, Regenstein MJ, Ren J (2011) In vitro antioxidant activity and in vivo anti-fatigue effect of loach (Misgurnus anguillicaudatus) peptides prepared by papain digestion. Food Chem 124: 188-194
- You L, Zhao M, Cui C, Zhao H, Yang B (2009) Effect of degree of hydrolysis on the antioxidant activity of loach (Misgurnus anguillicaudatus) protein hydrolysates. Innov Food Sci Emerg 10: 235-240
- Shevchenko A, Tomas H, Havliš J, Olsen VJ, Mann M (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856-2860.
- Encinar RJ, Ouerdane L, Buchmann W, Tortajada J, Lobinski R, et al. (2003) Identification of water-soluble selenium-containing proteins in selenized yeast by size-exclusion-reversed-phase HPLC/ICPMS followed by MALDI-TOF and electrospray Q-TOF mass spectrometry. Anal Chem 75: 3765-3774.
- Fiaschi T, Cozzi G, Raugei G, Formigli L, Ramponi G, et al. (2006) Redox regulation of beta-actin during integrin-mediated cell adhesion. J Biol Chem 281: 22983-22991
- Johansson M, Lundberg M (2007) Glutathionylation of beta-actin via a cysteinyl sulfenic acid intermediary. BMC Biochem 8: 26
- Metes-Kosik N, Luptak I, DiBello MP, Handy ED, Tang SS, et al. (2012) Both selenium deficiency and modest selenium supplementation lead to myocardial fibrosis in mice via effects on redox-methylation balance. Mol Nutr Food Res 56: 1812-1824
- Arthur JR (1988) Effects of selenium and vitamin E status on plasma creatine kinase activity in calves. J Nutr 118: 747-755.
- Kaur P, Bansal MP (2004) Influence of selenium induced oxidative stress on spermatogenesis and lactate dehydrogenase-X in mice testis. Asian J Androl 6: 227-232.
- 25. Maines MD, Kappas A (1976) Selenium regulation of hepatic heme metabolism: Induction of delta-aminolevulinate synthase and heme oxygenase. P Natl Acad Sci USA73: 4428-4431.
- . Maines MD, Kappas A (1976) Selenium regulation of hepatic heme metabolism: Induction of delta-aminolevulinate synthase and heme oxygenase. P Natl Acad Sci USA73: 4428-4431
- Hsieh Y, Jiang S (2013) Determination of selenium compounds in food supplements using reversed-phase liquid chromatography-inductively coupled plasma mass spectrometry. Microchem J. 110: 1-7
- Capelo JL, Ximenez-Embun P, Madrid-Albarran C, Camara C (2004) Enzymatic probe sonication: Enhancement of protease-catalyzed hydrolysis of selenium bound to proteins in yeast. Anal Chem 76: 233-237
- Ballihaut G, Pécheyran C, Mounicou S, PreudâÂ?Â?homme H, Grimaud R, et al. (2007) Multimode detection (LA-ICP-MS, MALDI-MS and nanoHPLC-ESI-MS2) in 1D and 2D gel electrophoresis for selenium-containing proteins. TRAC-Trend Anal Chem 26: 183-190
- Chitta RK, Landero-Figueroa AJ, Kodali P, Caruso AJ, Merino JE (2013) Identification of selenium-containing proteins in HEK 293 kidney cells using multiple chromatographies, LC-ICPMS and nano-LC-ESIMS. Talanta 114: 25-31
- Giusti P, Schaumlöffel D, PreudâÂ?Â?homme H, Szpunar J, Lobinski R (2006) Selenopeptide mapping in a selenium-yeast protein digest by parallel nanoHPLC-ICP-MS and nanoHPLC-electrospray-MS/MS after on-line preconcentration. J Anal Atom Spectrom 21: 26-32
- Zhang S,Wang Y, Bu D, Zhang H, Sun S (2011) ProbPS: A new model for peak selection based on quantifying the dependence of the existence of derivative peaks on primary ion intensity. BMC Bioinformatics12:346-355
- Gammelgaard B, Gabel-Jensen C, Stürup S, Hansen RH (2008) Complementary use of molecular and element-specific mass spectrometry for identification of selenium compounds related to human selenium metabolism. Anal Bioanal Chem 390: 1691-1706
- Jayasinghe BS, Caruso AJ (2011) Investigation of Se-containing proteins in Bertholletia excelsa H.B.K (Brazil nuts) by ICPMS, MALDI-MS and LC-ESI-MS methods. Int J Mass Spectrom 307: 16-27
- Sun S, Yang F, Yang Q, Zhang H, Wang Y, et al. (2011) Ms-Simulator: Predicting y-ion intensities for peptides with two charges based on the intensity ratio of neighboring ions. J Proteome Res 11: 4509-4516
- Encinar RJ, Å liwka-Kaszynska M, Polatajko A, Vacchina V, Szpunar J (2003) Methodological advances for selenium speciation analysis in yeast. Anal Chim Acta 500: 171-183
Relevant Topics
- Analytical Biochemistry
- Applied Biochemistry
- Carbohydrate Biochemistry
- Cellular Biochemistry
- Clinical_Biochemistry
- Comparative Biochemistry
- Environmental Biochemistry
- Forensic Biochemistry
- Lipid Biochemistry
- Medical_Biochemistry
- Metabolomics
- Nutritional Biochemistry
- Pesticide Biochemistry
- Process Biochemistry
- Protein_Biochemistry
- Single-Cell Biochemistry
- Soil_Biochemistry
Recommended Journals
- Biosensor Journals
- Cellular Biology Journal
- Journal of Biochemistry and Microbial Toxicology
- Journal of Biochemistry and Cell Biology
- Journal of Biological and Medical Sciences
- Journal of Cell Biology & Immunology
- Journal of Cellular and Molecular Pharmacology
- Journal of Chemical Biology & Therapeutics
- Journal of Phytochemicistry And Biochemistry
Article Tools
Article Usage
- Total views: 3642
- [From(publication date):
June-2017 - Apr 06, 2025] - Breakdown by view type
- HTML page views : 2758
- PDF downloads : 884