Promoter Analysis of a Powdery Mildew Induced Vitis vinifera NAC Transcription Factor Gene
Received: 27-Dec-2016 / Accepted Date: 09-Jan-2017 / Published Date: 16-Jan-2017 DOI: 10.4172/2329-8863.1000252
Abstract
Grapevine is an economically important crop throughout the world. However commonly cultivated species Vitis vinifera is susceptible to powdery mildew fungi. The objective of this study was to analyze the promoter of VvNAC36 transcription factor gene, which was induced by powdery mildew infection, as it was shown by previous microarray results. Analysis of VvNAC36 transcription factor promoter allows identifying the important cis-elements, which are responsible for gene regulation during infection. Knowing that the responsible cis-element of the gene is crucial for further study, the role of this gene is in powdery mildew infected grapevine. The study was performed using circumspect methodology. To confirm the presence of transgene in transgenic plants the PCR and glufosinate total herbicide application was used. The transplanted transgenic seedlings were infected with Golovinomyces orontii. To detect the GUS reporter gene expression, histochemical GUS staining and spectrophotometric assays were applied in each deletion variant. Finally the promoter sequence was analyzed by the PLACE (a database for PLAntCisacting Elements) program for identifying cis-elements. The results of the study showed significant difference in mock treated and induced expression in transgenic plants transformed with VvNAC36 promoter segments of different lengths, fused to GUS reporter gene. In basal expression level, the deletion variants 1178 bp and 257 bp differed significantly. Powdery mildew infection significantly increased the expression in all promoter variants, except the two shortest fragments. Based on the promoter motif analysis, it can be concluded, that the GT1-box (GAAAAA) supposed to have crucial role in regulating of VvNAC36 transcription factor gene during powdery mildew infection.
Keywords: VvNAC36 transcription factor; Powdery mildew; Gene expression; Cis-elements
405286Introduction
Grapevine (Vitis vinifera) is one of the oldest plants in the world (65 million years ago) [1]. It is an important fruit crop to produce wine, jam, juice, jelly, raisins, vinegar, grape seed extracts and oil. The wine sector is estimated to contribute more than 51.8 billion dollars to the US economy [2]. However the most grapevine growing country uses Vitis vinifera varieties, which are liable for best quality, but are vulnerable to powdery mildew fungus. Powdery mildew (PM) fungi are widespread and ever present among crop plants [3]. The reduction of yield due to PM each year probably exceeds the reduced yield, caused by any other plant disease. These pathogens rarely kill their hosts, but due to the utilization of their nutrients it lead to, a decrease in photosynthesis, increase of respiration and transpiration, impair me of growth and finally the plant produces 20-40% diminished yield [4].
Powdery mildew is controlled by systemic fungicides application. Beside that these chemical treatments lead to develop fungicide resistant mutants, which are superadded to the environmental pollution too. This implies that chemical treatment is not the best conceivable controlling method. Therefore elucidating the molecular level of plant-pathogen, interaction would approximate the background of resistance.
A large part of plant genes code transcription factors. In Arabidopsis ~2500 genes code transcription factors [5]. Transcription factors are the members of multigene families. The NAC transcription factor gene family is well studied. NAC transcription factor (no apical meristem growth gene, Arabidopsis activation factor gene, cup-shaped cotyledon response gene) genes are present only in plants [1,6]. The N-terminal region of the protein is divided into five conserved region and is used in DNA binding and dimerization. The C-terminal region has variable sequence and is involved in transcription activation or repression.
In addition, common feature of NAC protein C-terminal regions is characterized by the occurrence of simple amino acid repeats such as serine, threonine, proline and glutamine, or acidic residues [7]. The regulation of NAC transcription factors depend on the environmental condition. Approximately 20 to 25% of NAC transcription factor genes are expressed in response to one type of abiotic or biotic stress [5]. NAC transcription factor has excessive role in plant metabolic activity during any change to transcribe different genes, which are responsible for that influential factor.
In Arabidopsis and rice NAC, genes have been reported to regulate programmed cell death in xylem, induction of flowering, leaf senescence, xylary fiber development root and shoot apical meristem development. Additionally, NAC transcription factors have important role in abiotic and biotic stress responses. ATAF1 was found to negatively regulate the defense reactions against necrotrophic fungal pathogens [1]. Over expression of OsNAC gene increased drought resistance in rice [8,9]. Most NAC gene in their promoter region conserved with hormonal and fungal elicitor responsive motifs [10,11]. Fourteen Chitin-responsive transcription factor genes as part of the plant defense reaction were significantly up regulated after chitin treatment in Arabidopsis [12]. HBOH13 and HBOH14 transcription factor genes were induced upon infection with the powdery mildew during early stages of infection in rubber tree (Hevea brasiliensis) through mRNA Differential Display [13].
VvNAC transcription facto genes have different roles in grape vine like tissue development, biotic and abiotic response. For instance VvNAC18, VvNAC29, VvNAC37, VvNAC65 and VvNAC70 were expressed at high levels at inflorescence; VvNAC26 plays key roles in response to abiotic stress; in biotic stress response 7 VvNAC increased expression after virus-infection and 10 were up-regulated after Bois noir infection [1]. In addition 7 VvNAC transcription factor genes have been up regulated by powdery mildew elicitor at different levels in the given hour post infection (phi) [14]. Among these 7 transcription factor genes VvNAC36 has been significantly up regulated by powdery mildew infection.
There are numerous studies in relation to genome-wide identification researched NAC domain genes in plants, of which the whole genome sequences were known. The genomes of Arabidopsis thaliana (117), Populustrichocarpa (120), Glycine max (152), Oryza sativa (151), grapevine (79) and banana (167) contain numerous NAC genes respectively [5,15]. The frequency of transcription factor indicates the genome multiplication. For example V. vinifera genome, which has 79 NAC genes, changes more gradually than other plant genomes.
Materials and Methods
In the experiment the putative full length NAC promoter (3900 bp), four deletion variants (2935, 2456, 1178, 257 bp) and a promoterless construct were tested for induction by powdery mildew with three replication. These promoter fragments were fused to GUS reporter gene and each of them was transferred into wild type Arabidopsis thaliana by Gateway binary vector pGWB633 DNA. The selection marker was glufosinate total herbicide resistance ‘bar’ gene in the transgenic plants. For the evaluation the following methodology was investigated.
Transgenic line selection with glufosinate
The third generation (T3) independent lines transgenic Arabidopsis thaliana seeds were sown on autoclaved soil (Compo Sana). The two week old seedlings were sprayed by glufosinate containing total herbicide and due to the glufosinate resistance gene (bar gene), the positive transformants survived the application. Subsequently one week after application of herbicide, seedlings show clear selection, the positive transformants grow well, the plants, which did not contain the transgene, yellowed and died. Based on the Mendelian law of segregation the lines that contained 75% of surviving and 25% dying plants were selected for further analysis. Finally the surviving seedlings of these lines were transplanted into pots. The experiment had three replications of each type of transgenic plants.
Confirmation of transgene presence using PCR
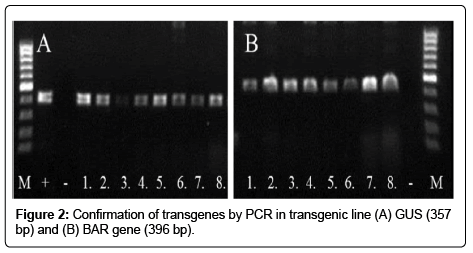
In addition to glufosinate application the integration of transgene was confirmed by PCR method. The DNA was extracted with a quick DNA preparation protocol [16]. Three transgenic plant leaves were placed into microcentrifuge tube and after adding 400 μl of extraction buffer (200 mM Tris-Cl (pH 7.5), 250 mM NaCl, 25 mM EDTA, 0.5% SDS) were ground with a micropestle (Figures 1 and 2).
The samples were centrifuged at 13000 rpm for 5 min. Subsequently 300 μl of the supernatant was transferred into a clean tube, 300 μl of 2-propanol was added and mixed by shaking. The mixture was centrifuged at maximum speed for 5 min and the supernatant was discarded carefully. The pellet was rinsed with 70% ethanol, drained, dried then dissolved in 100 μl of distilled water. 2.0 μl was used for the PCR reaction from the isolated DNA. For PCR reaction 1 μl WestTeam DNA polymerase buffer (10 ×), 1 μl dNTP (2 mM), 0.5 μl forward primer (10 μM), 0.5 μl reverse primer (10 μM), 0.25 μl WestTeam DNA polymerase enzyme (1 U/μl), 0.2 μl MgCl2 (20 mM) and 6.05 μl distilled sterile water was mixed. Both transgenes, the bar selection marker and NAC promoter: GUS fusion were tested by PCR applying the following primers: 5’AAACCCACGTCATGCCAGTT-3’ (BAR gene forward) and 5‘AAGCACGGTCAACTTCCGTA-3’ (BAR gene reverse) and 5’CCTAAAGCTTGGACAAACAGGAG3’ (NAC forward primer), 5’GCCCAACCTTTCGGTATAAAGAC-3’ (NAC reverse primer). Fragments were amplified using an initial denaturation step at 94°C for 2 min was followed by 30 cycles of 94°C for 35 s, 60°C for 30 s and 72°C for 45 s; finally followed by a final extension at 72°C for 15 min.
Powdery mildew infection
Three weeks after transplantation mock treatment and powdery mildew infection was applied on transgenic plants. For infection Golovinomyces orontii infected tobacco leaves were collected, the spores were washed off in 100 ml distilled water. This solution was sprayed on transgenic plants. For mock treatment healthy tobacco leaves were collected and washed in 100 ml distilled water. The infected and the mock treated transgenic Arabidopsis thaliana plants were maintained in a growth chamber at 22°C to 24°C in a 16 h light cycle to allow systemic infection.
GUS staining and microscopy observation
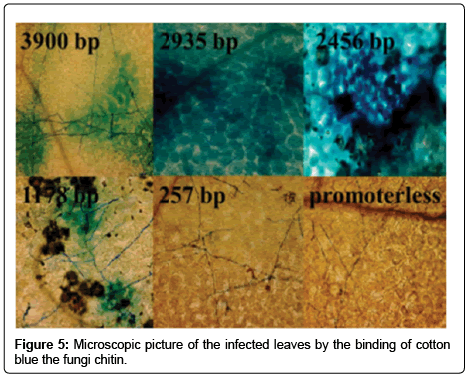
Fifteen days post infection the leaves were harvested for histochemical GUS assay. The infected leaves were placed into microcentrifuge tube and sufficient assay solution was added to cover them. To 1 ml of assay solution 10 μl of 30% hydrogen peroxide (H2O2) and 10 μl X-Gluc (50 mg/ml) solution was pipetted to them. The leaf samples were incubated overnight at 37°C. Finally 70% ethanol was added to the samples for the removal of chlorophyll from the tissues. The pictures were taken of these stained leaves. To confirm the GUS expression at the infected area microscopic pictures were taken from each deletion variant. To stain the fungus the chitin coloring cotton blue stain was used.
Spectrophotometric assay
Infected leaves were collected for spectrophotometric assay and placed immediately into liquid nitrogen. The frozen plant leaves were ground in GUS extraction buffer (50 mM Na-phosphatebuffer pH: 7.0, 10 mM β-mercaptoethanol and 0.1% triton-X 100; 1 μl extraction buffer/1 mg plant material). The homogenate was centrifuged in a microcentrifuge at 12,000 rpm for 10 min at 4°C. One mM p-nitrophenylβ-D-glucuronide (PNPG Sigma) substrate (final concentration) was added to the supernatant and it was incubated overnight at 37°C. The reaction was stopped by freezing the samples at -20°C. The GUS enzyme activity was measured at absorbance of 405 nm using Nano Drop 1000 spectrophotometer. The promoter-less samples were used as blank for both mock and infected samples. The absorbance of p-nitrophenol was measured according to Gilmartin and Bowler [17] and applied for estimating GUS expression.
Cis-element search of VvNAC promoter
The sequenced 3900 bp long VvNAC36 transcription factor promoter was analyzed by database of PLAntCis-acting Elements (PLACE). The inducible and repressor regulatory cis-elements were identified in the sequence at the website of http://www.dna.affrc.go.jp [18].
Statistical analysis
To analyze the quantitative spectrophotometric data the powerful and comprehensive data analyzing R-commander software was applied [19].
Results
Selection of transgenic Arabidopsis plants
The selections of herbicide resistant seedlings after glufosinate spraying were identified. Surviving green planlets were further analyzed by PCR to prove the integration of both the bar and NAC promoter sequences.
Promoter sequence analysis
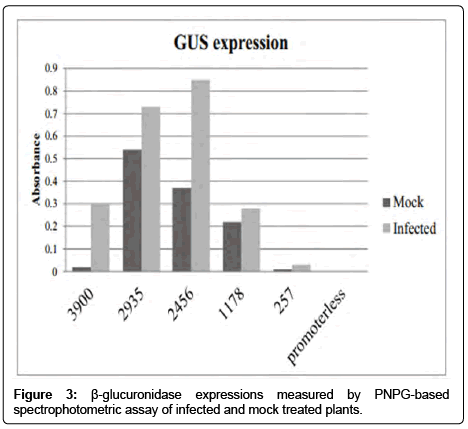
The results of the GUS activity was driven by the longest VvNAC promoter, four deletion, 2935 bp (removed 965 bp from full length), 2456 bp (removed 1444 bp from full length), 1178 bp (removed 2722 bp from full length) and 257 bp (removed 3643 bp from full length) and a promoter-less construct on the effect of mock treatment and powdery mildew infection are shown in Figure 3.
Promoter deletion activity analysis (only in mock)
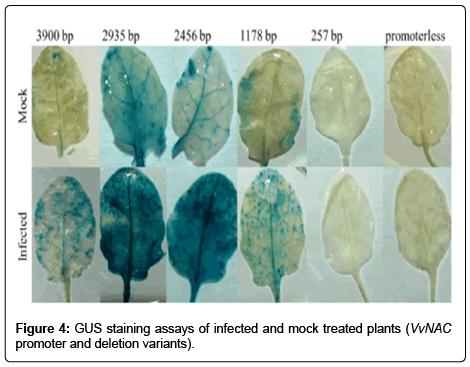
The spectrophotometrically measured GUS reporter gene expression of 2935, 2456 and 1178 bp promoters did not differ significantly in the mock treatments. However the full length promoter 3900 bp showed significantly lower expression than the above three deletion variants (Figure 3). Likewise the plant carrying the 257 bp promoter, the GUS expression showed 22 times lower expression than deletion variant 1178 bp. Beside the spectrophotometric data the histochemical GUS staining assay indicates the same expression levels; significant differences were observed in full length and 257 bp promoter carrying plants compared to the other deletion variants (Figure 4). These data show that the 1178 bp to 2935 bp long NAC promoters activate GUS expression even without PM infection (Table 1).
| Df | Sum Sq | Mean Sq | F value | Pr(>F) | |
|---|---|---|---|---|---|
| Deletion variant | 5 | 0.7549 | 0.15098 | 3.359 | 0.0397* |
| Residuals | 12 | 0.5393 | 0.04495 |
S2=0.173
Table 1: ANOVA summary of PNP absorbance data of mock treated plants.
Promoter deletion activity analysis after PM infection
On the effect of powdery mildew infection the GUS expression increased in all five variants (Table 2). The highest level was achieved in VvNAC36 promoter deletion variants 2935 bp and 2456 bp, the difference between these two ones was not significant. In contrast the plant carrying 3900 bp (full length VvNAC36 promoter), 1178 bp and 257 bp VvNAC36 promoter displayed significantly lower expression compared to the plants carrying the 2935 bp and 2456 bp long promoter sequence (Figures 3 and 4).
| Df | Sum Sq | Mean Sq | F value | Pr(>F) | |
|---|---|---|---|---|---|
| Deletion variant | 5 | 1.8972 | 0.3794 | 14.21 | 0.000109*** |
| Residuals | 12 | 0.3204 | 0.0267 |
S2=0.133
Table 2: ANOVA summary of PNP absorbance data of infected plants.
The longest (3900 bp promoter showed a 15 fold change in the response to infection. The 2456 bp deletion variant changed 2.3 times 2935 bp and.1178 bp deletion variants which increased 1.3 times the expression of GUS due to the pathogen. Besides, the difference between gene regulations of mock and infected treatment is the variation which was observed within each respective pathogen which induces the expression of deletion variants too. Comparing the 1178 bp and 257 bp deletion variant to 2456 bp promoter a 3 fold (1178 bp) and 27 fold (approaching zero expression, 257 bp) reduce was observed in the regulation of GUS respectively. However the induced regulation of 3900 bp and 1178 bp deletion variants showed almost the same GUS expression.
The pictures of GUS staining assay confirm the results of spectrophotometry, where the reporter gene expression level was influenced by the deletion variants (Figure 4). The infected area of 3900 bp, 2935 bp, and 2456 bp promoter containing leaves showed high GUS expression in the microscopic photos (Figure 5). In contrast the infected leaves of plants containing 1178 bp VvNAC36 promoter did not show high increase of GUS expression in the infected area. Unlike the VvNAC36 257 promoter length that showed a lack of GUS expression in the infected area of leaves.
Putative cis-elements responsive to inducer or repressor agent
Using the plant Cis-acting elements, plant promoter analysis program at http://wwwdna.affrc.go.jp several cis-elements were identified in VvNAC36 transcription factor promoter, which response to inducer or repressor (silencer) agent. Table 3 details the identified cis-elements with the respective inducer or repressor agent and location on strands (+/-).
| Cis-element | Inducer | Location of cis -element on both Strand |
|---|---|---|
| TATTCT | Blue, white or UV-A light | 2168(-) |
| ACGTG | Dehydration stress or dark. | 1732 (+) and 2781 (+) |
| TACGTA | Sugar-repression | 2553 (+) and 2553 (-) |
| ACGT | Dehydration stress or dark-. | 2553 (-), 1732 (+),2554 (+), 2781 (+), 2884(+), 1732 (-), 2554 (-), 2781(-) and 2884(-) |
| TATCCAT | Sugar starvation | 3381(-) and 3401(-) |
| AAACAAA | Anaerobically | 569(+), 3365(+), 2128 (-), 2440 (-), and 3429(-) |
| NGATT | Cytokinin | 2173 (+), 1231 (+), 2675 (+), 2877 (+), 553 (+), 815 (+), 1687 (+), 1709 (+), 1863 (+) , 2217 (+), 116 (+), 1494 (+), 1598 (+),1721 (+), 1982 (+), 2063 (+), 2679 (+),3020 (+), 3079 (+), 3182 (+), 3581 (+), 3890 (+), 127 (-), 395 (-), 465 (-), 528 (-), 712 (-), 1128 (-),1384 (-), 2038 (-), 2460 (-), 2580 (-), 2616 (-), 2656 (-), 3114 (-), 3446 (-), 3461 (-)3745 (-) |
| TGACG | Auxin and/or salicylic acid | 270 (+),1735 (+), 2779 (+) and 2603 (-) |
| ACCWWCC | Elicitor treatment or UV-B irradiation | 2525 (+) |
| TTGACC | Pathogen elicitor | 3235 (+) , 99 (-), 959 (-) and 1698 (-) |
| AWTTCAAA | Ethylene | 2903 (+), 3410 (+),3518 (+), 493 (-), 1008 (-) and 2509 (-) |
| GCCGCC | Jasmonate or ethylene | 1293 (-) |
| GAAAAA | Pathogen-or salt | 442 (+), 1254 (+),1557 (+), 2308 (+),1539 (-), 1842 (-), 1887 (-), 2281 (-), 2682 (-), 3542 (-), 3584 (-),3673 (-),and 3715 (-) |
| CTCTT | Mycorrhizal | 5 (+), 11 (+), 1399 (+), 3359 (+),354 (-) , 446 (-), 734 (-) and 1081 (-) |
| TGTATATAT | Light-repressed | 2989 (+) and 513 (-) |
| TTATCC | Sugar-repressive | 1506 (-), 3383 (-) |
| AATACTAAT | Sucrose | 853 (+) |
| AACGTG | Jasmonate (JA) | 1731 (+) |
| TATCCAY | Sugar-repression | 3381 (-), and 3401 (-) |
| TTTGACY | Elicitor-induced | 3234 (+) ,1312 (-) and 1698 (-) |
| TTGAC | Salicylic acid (SA)-induced | 485 (+) , 754 (+), 1573 (+), 2101 (+), 2786 (+), 3235 (+), 3609 (+), 100 (-), 960 (-) 1313 (-),1699 (-), and 2604 (-) |
| CTGACY | Elicitor-induced | 2449 (-) and 3808 (-) |
Table 3: Identified cis-elements of VvNAC36 promoter region responsible in interaction with inducer or repressor. Green shading indicates repressor responsive motifs and yellow highlighting indicates pathogen-related motifs.
As the PLACE identified the VvNAC36 promoter it contains several inducible and some repressor responsive motifs. Among the inducible cis-elements, pathogen elicitor, salicylic acid (SA), cytokinin and dehydration or dark stress motifs were the most conserved elements in the promoter region. The GT-1 box (GAAAAA) and the W- box core motifs (TGAC) were conserved mostly among the pathogen elicitor induced cis-elements. The PLACE identified sugar and lightrepressive cis-element especially in the upstream region of the full length promoter. Two sugar repressive cis-elements were identified TATCCAY at 3381(-), 3401(-), and TTATCC at 1506 (-), 3383 (-), 2553 (+) and 2553 (-). Light repressive cis-element TGTATATAT was detected at 2989 (+) and 513 (-).
Discussion
Vitis vinfera NAC36 transcription factor gene was up regulated by powdery mildew pathogen. Based on the pathogen induced expression level of the longest (full length) promoter::GUS fusion in comparison to basal expression, it indicates that VvNAC36 transcription factor is a powdery mildew dependent gene and may be part of defense system. The basal level expression of GUS reporter gene in the full length fusion containing plants was significantly low (p-value=0.0256); the infection induced GUS reporter gene expression, showed a 15 fold increase. However when it was compared to the highly expressed 2456 bp deletion variant, it showed significantly 3 times less expression.
This result reveals that the full length promoter has some negative regulatory motifs in its sequence. Interestingly based on PLACE plant promoter analysis program, this region contains conserved TATCCAY and TTATCC sugar repressive cis-elements (Figure 6B). Mostly the plants have more sugars in their leaves due to photosynthesis metabolic activity. Therefore if the sugar repressive cis-element is conserved in this powdery mildew inducible promoter upstream region, the deletion of these repressive cis-elements causes a 3 fold up regulation of GUS reporter gene in 2456 bp deletion. Furthermore two sites of the full length promoter region contain light repressive cis-elements and these may contribute too, in the reduction of GUS regulation of full length promoter containing plants. Further investigations are necessary to experimentally confirm these assumptions.
Figure 6: Site of W and GT1 boxes on NAC promoter (A) Original sequence of 2456 bp NAC promoter; W and GT1 boxes, 2456 bp, 1178 bp, 257 bp primer sequences are indicated (B) Schematic diagram of location of responsive cis-elements of VvNAC deletion variants and GUS reporter gene fusions: Δ W-box core motif (TGAC), * Elicitor responsive GT1-boxes (GAAAAA) and Ο Sugar repressive cis-element.
In addition, the second longest promoter (2935 bp) which contains sugar repressive TACGTA cis-elements at 2553 (+) and at 2553 (-) position and showed a little bit lower expression than the 2456 bp promoter fusion in infected treatment, but the difference was not significant. This motif may cause reduction of GUS reporter gene expression by blocking the elicitor responsive inducer. However the induction of basal expression by this promoter was higher than in 2456 bp fusion. This indicates that the repressor site may not influence the general transcription factor protein induced basal expression. In mock treated plant, the full length promoter::GUS reporter gene fusion showed 27 times less GUS expression than the deletion variant 2935 bp probably due to the presence of these sugar and light repressive ciselements.
Therefore this result revealed that the light and sugar repressive binding cis-elements cause a significantly low inducement of GUS reporter gene by full length VvNAC36 promoter in both mock and infected treatment. However in this promoter::GUS, fusion showed 15 times higher up regulation in response to infection compared to mock treatment, which confirms that VvNAC36 is a powdery mildew inducible gene.
The basal expression of the reporter genes in plants carrying 1178 VvNAC36 promoter was significantly 22 times fold than the 257 bp fragment containing plants. Evaluation of GUS activity in mock treated plants carrying 1178 bp and 257 bp deletion constructs indicates that reduction of the 1178 bp length VvNAC36 promoter to 257 bp results significantly decrease in expression. This showed that positive regulatory cis-element is apparently located between 1178 to 257 bp in the promoter region, and this is responsible for basal expression. The powdery mildew induced GUS reporter gene expression significantly differ in 2456 bp and in 1178 bp VvNAC36 promoter construct meaning that elicitor responsive motifs may be found in 2456 to 1178 bp region. Two putative regulatory motifs, pathogen elicitor responsive cis-acting elements were identified in the promoter region of the VvNAC36 gene. These cis-elements, W-boxes (TGAC) and GT1-boxes (GAAAAA), are considered being pathogen elicitor responsive in plants. However, based on the detailed evaluation the promoter region of 2456 bp was highly significant from 1178 bp and 257 bp VvNAC36 promoter deletion variant respectively (Figure 6A).
In the VvNAC36 2456 bp promoter region fifteen W-boxes and eight GT1-boxes were detected on both strands (Figure 6A). Seven W-boxes out of the total fifteen boxes and seven GT1-boxes of the total eight boxes were dropped out from VvNAC36 1178 bp promoter sequence. The 1178 bp VvNAC36 deletion variant promoter::GUS reporter gene fusion showed significantly low expression when compared to the VvNAC36 2456 bp promoter construct due to the deletion of seven W-boxes and seven GT1-boxes. In addition the total eight GT1-boxes and fourteen W-boxes were absent in VvNAC36 257 promoter regions, the single W-box is not able to regulate neither the basal and induced expression of 257 bp promoter::GUS fusion. The 257 bpVvNAC36 deletion variant promoter::GUS reporter gene fusion showed a large decrease in expression due to the totally lost GT1-boxes and only a single W-box presence.
In this study the VvNAC36 transcription factor promoter was analyzed to identify the motif(s), which is/are responsible for up regulation of the gene in powdery mildew infection. In infected treatment among six transgenic lines including full length promoter and promoter less construction, the promoter length of 1178 bp and 257 bp showed significantly low GUS gene expression. In the mock treatment the reporter gene expression was significantly low only in the plant transformed with the 257 bp deletion variant. The results suggest that putative motifs found between 1178 bp to 2456 bp deletion variant are critical for powdery mildew elicitor induction and is between 1178 bp to 257 bp deletion variant. The core motif found in the sequence is required for basal expression regulation of the VvNAC36 transcription factor gene.
The detected responsible motifs found in 2456 bp and 1178 bp deletion variants were putative motifs depending on powdery mildew elicitor. A total fifteen core W-boxes (TGAC) and eight GT1-boxes (GAAAAA) were identified in the promoter of 2456 bp length while there were only eight W-boxes (TGAC) and one GT1- box in the 1178 bp promoter region. These pathogen responsive motifs have outstanding role in up regulation in the gene by pathogen elicitor. For example, in soybean suspension-culture cells the expression of SCaM-4 gene, which contains GT1-boxes in its promoter, was dramatically induced within 1 h after treatment with Pseudomonas syringaepvglycinea P. syringaepv, while the application of exogenous hydrogen peroxide, salicylic acid, Jasmonic acid did not increase the transcription of this gene. However mutations in the GT1-boxes significantly reduced the expression of this gene [20]. W-boxes are the major elicitor responsive motifs in most pathogen related protein coding genes [21].
The GT1 and W-boxes motifs in Vitis vinifera NAC36 transcription factor promoter play important role for expression regulation in powdery mildew infection especially GT1-boxes, which occurs only once in promoter length 1178 and are totally absent in the 257 deletion variant, involve in the responsible transcription factor binding site. Total 8 W-boxes, only one GT1-box was detected in the 1178 deletion variant. Therefore based on the significant expression drop between 2456 bp and 1178 bp promoters the conclusion can be drawn that, the GT1 box has an important regulation role in the PM induced gene expression of VvNAC36 transcription factor gene. The powdery mildew regulated VvNAC36 transcription factor gene may have positive role in defense system against powdery mildew infection. As the previous microarray study indicated, the high expression of VvNAC36 transcription factor gene was only regulated by powdery mildew and not by exogenous salicylic acid treatment [14]. This study identified the responsible motif for regulation of this gene, the GT-1 box, which is regulated by elicitor not by salicylic acid.
Therefore it would be worthwhile examining, how the mutation in GT1 box of VvNAC36 promoter influences the expression on the effect of powdery mildew infection and to study the functional role of VvNAC36 transcription factor gene using complementation test.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Acknowledgements
First I would like to give my great acknowledgement to GOD, who never aparts me so far and forever in each my activity for this work too. Secondly I would like to thank Zsofia Toth for her great assist in both laboratory experiments and in editing this work. Many thank for her general consultation to Dr. Erzsebet Kiss too. I appreciate the support of FAO and Hungarian government.
References
- Wang N, Zheng Y, Xin H, Fang L, Li S (2012) Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Reports 32: 61-75.
- Broome J, Warner C, Keith D (2008) Agro-environmental partnerships facilitate sustainable wine-grape production and assessment. California Agriculture 62: 133-141.
- USEPA (2005) Use of genetic toxicology information for risk assessment. USEPA-US.
- Agrios GN (2005) Plant Pathology. Elsevier Academic Press, Amsterdam, p: 952.
- Puranik S, Sahu PP, Srivastava PS, Prasad M (2012) NAC proteins: regulation and role in stress tolerance. Plant Science 17: 369-381.Â
- Kikuchi K, Ueguchi TM, Yoshida KT (2001) Molecular analysis of the NAC gene family in rice. Molecular and General Genetics 262: 1047-1051.
- Olsen AN, Ernst HA, Lo LL, Skriver S (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends in Plant Science 10: 79-87.
- Carviel JL, Al-Daoud F, Neumann M, Mohammad A, Provart NJ, et al. (2009) Forward and reverse genetics to identify genes involved in the age-related resistance response in Arabidopsis thaliana. Molecular Plant Pathology 10: 621-634.
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2005) The NAC Transcription Factors NST1 and NST2 of Arabidopsis Regulate Secondary Wall Thickenings and Are Required for Anther Dehiscence the Plant Cell. American Society of Plant Biologists 17: 2993-3006.
- Jensen MK, Kjaersgaard T, Petersen K, Skriver K (2010) Time-specific regulators of hormonal signaling in Arabidopsis. Plant Signaling and Behavior 5: 907-910.
- Fujita M, Yasunari F, Yoshiter N, Takahashi F, NarusakaY, et al. (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Current Opinion in Plant Biology 9: 436-442.
- Libault M, Wan J, Czechowski T, Udvardi M, Stacey G (2007) Identification of 118 arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol Plant-Microbe Interact 20: 900-911.
- Li X, Zhenghong B, Rong D, Peng L, Qiguang H, et al.(2016) Identification of Powdery Mildew Responsive Genes in Hevea brasiliensis through mRNA Differential Display. International Journal of Molecular Sciences 17: 181.
- Fung RWM, Martin G, Csaba F, Laszlo GK, Yan H, et al. (2008) Powdery mildew induces defense-oriented reprogramming of the transcriptome in a susceptible but not in a resistant grapevine. Plant Physiol 146: 236-249.
- Cenci A, Guignon V, Roux N, Rouard M (2014) Genomic analysis of NAC transcription factors in banana (Musa acuminata) and definition of NAC orthologous groups for monocots and dicots. Plant Molecular Biology 85: 63-80.
- Glazebrook J (2002) Arabidopsis a Laboratory Manual. Cold Spring Harbor Laboratory Press, New York.
- Gilmartin PM, Bowler C (2002) Molecular Plant Biology. Oxford University Press, London, UK.
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Research 27: 297-300.
- Fox J, Weisberg S (2011) An R and S-plus companion to Applied Regression. Sage, India.Â
- Park HC, Kim ML, Kang YH, Jeon JM, Yoo JH, et al. (2004) Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant physiology 135: 2150-2161.
- Eulgem T (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Science 10: 71-78.
Citation: Gebrie SA (2017) Promoter Analysis of a Powdery Mildew Induced Vitis vinifera NAC Transcription Factor Gene. Adv Crop Sci Tech 5:252. DOI: 10.4172/2329-8863.1000252
Copyright: © 2017 Gebrie SA. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 4946
- [From(publication date): 0-2017 - Jul 04, 2025]
- Breakdown by view type
- HTML page views: 4027
- PDF downloads: 919