Opinion Article Open Access
Probing Potent Interfacial Residues in Microvirin before Complexation with Glycoproteins: Visualizing Anti-HIV Activity in silico
Paramasivan Manivannan and Gangatharan Muralitharan*Department of Microbiology, Bharathidasan University, Tiruchirappalli, India
- Corresponding Author:
- Gangatharan Muralitharan
Department of Microbiology
Bharathidasan University
Tiruchirappalli, Tamilnadu, India
Tel: +91 431 2407082
E-mail: drgm@bdu.ac.in
Received Date: March 02, 2016; Accepted Date: October 04, 2017; Published Date: October 06, 2017
Citation: Manivannan P, Muralitharan G (2017) Probing Potent Interfacial Residues in Microvirin before Complexation with Glycoproteins: Visualizing Anti-HIV Activity in silico. J Marine Sci Res Dev 7:241. doi:10.4172/2155-9910.1000241
Copyright: © 2017 Manivannan P, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Marine Science: Research & Development
Abstract
Microvirin, a mannose binding anti-HIV (Human Immunodeficiency virus) lectin has been proved as a potent agent in combating HIV. Ligand binding residues of Microvirin was taken in to account for analysis of interfacial residues and identify surface/Buried residues based on Accessible surface area and free energy based calculations. After categorization, Comparative B factors were assessed for prediction of Rigid/Non-Rigid residues among the Ligand binding residues. Furthermore docking microvirin with glycoproteins gp120 and gp41 revealed the conserved interfacial residues in complexes and affirms the computational dissection of microvirin’s anti-HIV activity.
Keywords
Anti-HIV activity; B factor analysis; Interfacial residues; Ligand binding residues; Microvirin
Introduction
Acquired immune deficiency syndrome (AIDS) is a fatal disease of the human immune system caused by the human immunodeficiency virus (HIV). Human Immunodeficiency virus (HIV) is still a significant risk of high morbidity and mortality with secondary infections and devastated over 25 million deaths globally [1]. The disease is characterized by opportunistic infections and tumors, which render the individual’s immune system debilitating. HIV-1 entry in to host cell is mediated by viral envelope glycoproteins gp120 and gp41. Initially after binding to CD4 cells they bind on to CXCR4 and CCR5 [2,3]. Anti-HIV medication broadly fall in to three main categories namely nucleoside Reverse Transcriptase, Non- Nucleoside Reverse Transcriptase inhibitors and protease inhibitors. However, drugs presently in the pipeline are administered combinatorially for arresting the pathogen [4]. Nevertheless the genetic diversity of HIV virus still remains quizzical for several chemists and Biologists as Human Immunodeficiency virus evades the combating mechanism at ease. Vaccines for HIV have been still in testing stages which comprise of Subunit vaccines [5], Recombinant viral vaccines [6] and DNA vaccines [7]. But there has been reduced success in developing an anti- HIV vaccine. Cyanobacterial lectins like Cyanovirin-N, 11-kDa protein isolated from the cyanobacterium Nostoc ellipsosporum is effective against both laboratory strains and primary isolates of HIV-1 and HIV-2 at an order of nanomolar concentration. Cyanovirin-N aborts cell-to-cell fusion and transmission of HIV-1 infection [8]. Antiviral activity is attributed to the binding with the viral envelope glycoprotein gp120. Nevertheless the lectin microvirin (MVN) from the cyanobacterium Microcystis aeruginosa PCC7806 shows 33% identity at the amino acid level with CV-N. MVN has anti-HIV-1 activity comparable with that of CV-N but a much higher safety profile [9]. Microvirin (MVN) compared with cyanovirin-N (CVN), show potent HIV-1 neutralization with reduced toxicity profiles [10]. The present analysis deals with ligand binding properties of microvirin for categorizing surface and buried residues based upon solvent accessibility and free energy of ligand binding residues. Docked complex interfacial residues also affirm the fact that majority of interfacial properties in interacting residues.
Methods
Ligand binding site prediction
Ligand binding sites were predicted using Q site finder [11], surface topology and pocket information were analyzed by the castP server [12]. Pocket detection and occupancy was found using Q-SiteFinder. Clefts were identified in the protein-surface using Q-SiteFinder. The solvent accessible surface area (SASA) was found by GETAREA [13]. The atomic Solvent Accessible Surface Area (SASA) covered by each cleft was calculated by utilizing radius of water probe 1.4 A0 and the area/ energy per residue was calculated. Dielectric constant was set to 80.0, and Poisson-Boltzman method of computation for 20 cycles was used for calculating the electrostatic potential in SWISS-PDB viewer [14]. All the ligand binding residues were among hotspots as predicted by Meta-PPISP [15]. Furthermore, PIC [16] was made used to calculate the nature of interaction in the ligand binding residues.
Identification of surface/Buried residues
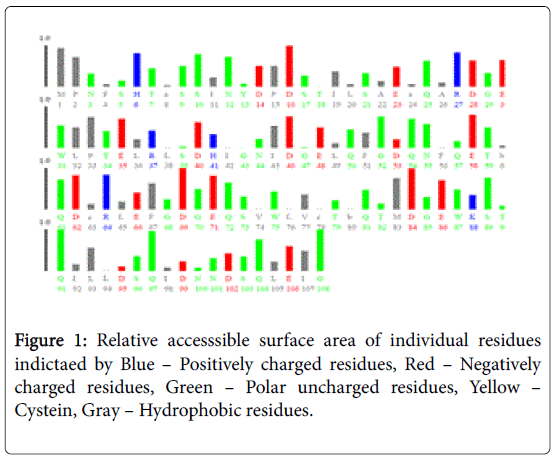
Two methods were adopted for presicting surface/buried residues among ligand binding residues. First, relative accessible surface area by Accessible solvent area calculations (ASAVIEW) [17] based calculations demarcated the residues based on five criteria namely positively charged, negatively charged, polar uncharged, cystein rich and hydrophobic residues. Among the predictions hydrophobic and positively charged residues were constituted as buried as amino acids are positively charged. Whereas the remaining criteria satisfied the surface residue pattern due to solvent accessibility. In total 18 residues were pooled as surface residues by this method. Secondly, GROMOS’96 version of SWISS-PDB viewer [14] was used to calculate G of individual ligand binding residue. Values of free energy in negative were designated as surface and positive values were regarded as buried residues as energies in Kcal/mol provide binding energies. Moreover, reduced binding energies yield high affinity.
Comparative B factor analysis and prediction of rigid/non rigid nature of ligand binding residues
Binding factor for ligand binding residues were predicted using BFPred [18] and individual B factors were statistically depicted for summarizing rigid/non rigid nature of the ligand binding residues. The normalized B-factor per residue (Bi,N) was calculated based on
Bi,N = Bi-<Bi>
σ Bi
where Bi - B-factor of residue
<Bi> - mean B factor
σ - Standard deviation
Bi,N ≤ 0.04 were considered as rigid whereas ≥ 0.04 were regarded non-rigid [19].
Results and Discussion
Ligand binding residues in a protein constitute a subset of hotspot residues in protein - protein interactions. Hotspot residues have been correlated to energy hot spots and structurally conserved residues. Hot spots in the interface of protein complexes identify unique binding sites from the rest of the surface [20]. Presently, microvirin a mannose binding lectin’s ligand binding residues were assessed for conservation and properties which renders them to be potential binders with glycoproteins of HIV. Table 1 depicts the ligand binding residues with the potent interaction whether they conform hydrophobic, catioinic-pi, ioinic and disulfide linkages. Tyr 13, Arg37, Cys 60,61 are responsible for Cationic – pi, ioinic and disulphide linkages respectively. Hotspot residues as predicted by meta-PPISP also affirm that ligand binding residues confer interfacial properties. Fourier transform MAP (FTMAP) scans also confer the hydrophobic and hydrogen bond interactions among ligand binding residues of microvirin (Table 2). The present approach combinatorially includes Kortemme and Baker with hydrogen bond contribution to hotspots and pertaining to hydrophobic interactions [21,22].
| Ligand binding residues | Type of interaction |
|---|---|
| Tyr 13 | Cationic-pi |
| Asp 14 | Hydrogen bond |
| Pro 15 | Hydrogen bond |
| Asp 16 | Hydrogen bond |
| Ser 17 | Hydrogen bond |
| Thr 18 | Hydrogen bond |
| Ile 19 | Hydrogen bond |
| Leu 20 | Hydrogen bond |
| Leu 36 | Hydrogen bond |
| Arg 37 | Ionic |
| Leu 38 | Hydrogen bond |
| Ser 39 | Hydrogen bond |
| Asp 40 | Hydrogen bond |
| Ile 42 | Hydrogen bond |
| Asn 55 | Hydrogen bond |
| Phe 56 | Hydrogen bond |
| Glu 57 | Hydrogen bond |
| Glu 58 | Hydrogen bond |
| Thr 59 | Hydrogen bond |
| Cys 60 | Disulfide bridge |
| Glu 61 | Hydrogen bond |
| Asp 62 | Hydrogen bond |
| Cys 63 | Disulfide bridge |
| Cys78 | Disulfide bridge |
Table 1: Interaction characteristics of ligand binding site residues of Microvirin.
| Ligand binding residues | Energy in Kcal/mol |
|---|---|
| Tyr 13 | -87.719 |
| Asp 14 | 11.07 |
| Pro 15 | -8.249 |
| Asp 16 | 22.317 |
| Ser 17 | 16.402 |
| Thr 18 | -21.202 |
| Ile 19 | -19.293 |
| Leu 20 | -36.652 |
| Leu 36 | -36.355 |
| Arg 37 | -227.821 |
| Leu 38 | -28.355 |
| Ser 39 | 19.881 |
| Asp 40 | 7.498 |
| Ile 42 | 9.271 |
| Asn 55 | -173.702 |
| Phe 56 | -50.602 |
| Glu 57 | -173.616 |
| Glu 58 | 11.874 |
| Thr 59 | -24.101 |
| Cys 60 | -16.145 |
| Glu 61 | -182.734 |
| Asp 62 | -2.57 |
| Cys 63 | 1.46 |
| Cys78 | -46.419 |
Table 2: Energy values of binding residues.
Rigid/Non rigid residues
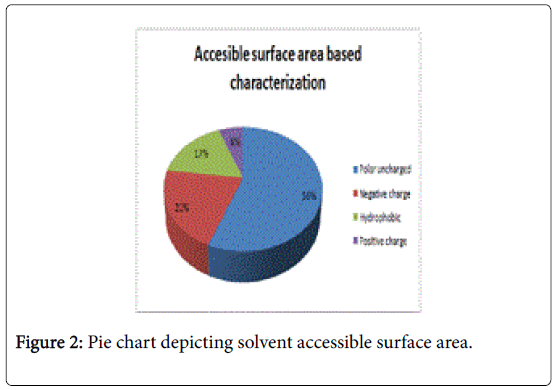
Figure 1 pictorially demonstrates individual nature of aminoacids in the lectin, microvirin. Free energy based calculations were done using GROMOS’96 version of SWISS-PDB Viewer. Negative delta G were regarded surface residues and positive values were considered buried in the present analaysis. Relative accessible surface area calculations revealed the presence of 18 surface residues (Figure 2) due to their polar uncharged and negative nature of aminoacids. As aminoacids are positively charged, hydrophobic and positively charged residues were regarded as buried.
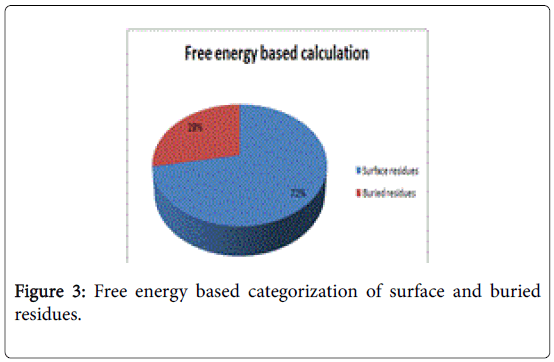
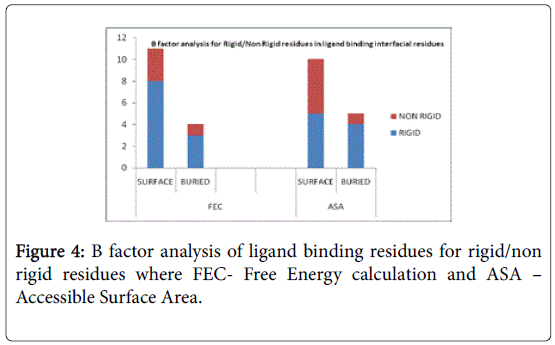
A total of 16 surface residues (Figure 3) were significant from the study. Although the energy based and surface based approaches revealed specific properties for ligand binding residues, Comparative B factor analysis (Figure 4) after B factor prediction by BFPRED server showed that rigid residues dominate non rigid residues in both relative accessible surface area and energy based calculations. In this context, self-protein stabilizing residues are alone taken into account for assessing their rigidity as the same method was used to analyze transient protein complexes [19] in which some interfacial residues stabilize the self-protein. Here a similar approach reveals that ligand binding residues apart from forming complexes do self-stabilize the protein. Hence the rigid/non rigid analysis yields predominant stability of ligand binding Residues.
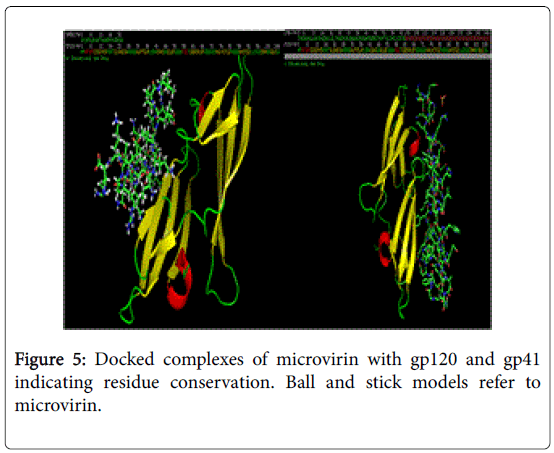
Conservation of ligand binding residues in docked complexes of microvirin with gp120 and gp41.
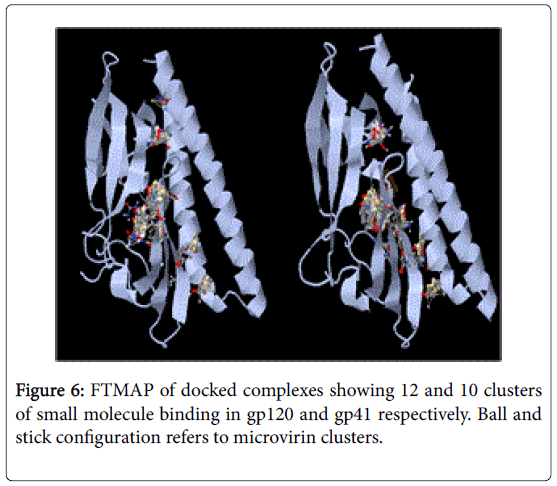
NFSHTFQE (Asn, Phe, Ser, His, Thr, Phe, Gln, Glu) were conserved in both docked complexes of glycoproteins with high binding affinities. Amongst which, His alone is not enlsited in ligand binding residue. However it may be a surface interacting residue (Figure 5). Structural and physicochemical signatures at Protein-protein interaction interfaces are enough to detect optimal ligand binding of unliganded protein [23]. Fourier transform mapping revealed 12 clusters of small molecule binding sites for gp120 interaction compared to 10 clusters with gp41 (Figure 6). Hence from the present study it is evident that ligand binding residues can act both as self-stabilizing and binding attributes. Furthermore it can be categorized as structural conservation of lectins like microvirin is effcieint in glycan binding with conservatory aminoacids.
Conclusion
Ligand binding residues between gp120 and gp41 show potent interaction consisting hydrophobic, catioinic-pi, ioinic and disulfide linkages. Tyr 13, Arg37, Cys 60,61 are responsible for Cationic – pi, ioinic and disulphide linkages. Rigid/non rigid analysis yields predominant stability of ligand binding Residues. NFSHTFQE (Asn, Phe, Ser, His, Thr, Phe, Gln, Glu) were conserved in both docked complexes of glycoproteins with high binding affinities. Fourier transform mapping revealed 12 clusters of small molecule binding sites for gp120 interaction compared to 10 clusters with gp41. Structural conservation observed from the present study of lectins like microvirin prove glycan binding is conserved in protein-protein interaction. Experimental data to prove the above phenomenon can be foreseen in the near future.
Acknowledgement
PM acknowledges University Grants Commission for award of UGC BSR meritorious Fellowship.
References
- Joint United Nations Program on HIV/AIDS (2008) Report on the Global AIDS Epidemic, UNAIDS, Geneva, Switzerland.
- Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, et al. (1990) Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J BiolChem 265: 10373-10382.
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, et al. (1998) Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 393: 648-659.
- Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, et al. (1998) The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 393: 705-711.
- Tandon VK, Chhor RB (2005) Current Status of Anti-HIV Agents. Curr.Med.Chem – Anti infective agents 4: 3-28.
- Heilman CA, Baltimore D (1998) HIV vaccines—where are we going? Nat. Med. 4: 532-534.
- Moldoveanu Z, Porter DC, Lu A, McPherson S, Morrow CD (1995) Immune responses induced by administration of encapsidated poliovirus replicons which express HIV-1 gag and envelope proteins. Vaccine 13: 1013-1022.
- Donnelly JJ, Ulmer JB, Shiver JW, Liu MA (1997) DNA vaccines. Annu Rev Immunol. 15: 617-648.
- Bewley CA, Gustafson KR, Boyd MR, Covell DG, Bax A, et al. (1998) Solution structure of cyanovirin-N, a potent HIV-inactivating protein. Nat Struct Biol. 5: 571-578.
- Huskens D, Ferir G, Vermeire K, Kehr JC, Balzarini J, et al. (2010) Microvirin, a novel alpha(1,2)-mannose-specific lectin isolated from Microcystisaeruginosa, has anti-HIV-1 activity comparable with that of cyanovirin-N but a much higher safety profile. J BiolChem 285: 24845-24854.
- Laurie AT, Jackson RM (2005) Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics 21: 1908-1916.
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, et al. (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucl. Acids Res 34: 116-118.
- Fraczkiewicz R, Braun W (1998) Exact and Efficient Analytical Calculation of the Accessible Surface Areas and Their Gradients for Macromolecules. J Comp Chem 19: 319-333.
- Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucl. Acids Res 33: 363-367.
- Qin SB, Zhou HX (2007) Meta-PPISP: a meta web server for protein-protein interaction site prediction. Bioinformatics 23: 3386-3387.
- Tina KG, Bhadra R, Srinivasan N (2007) PIC: Protein Interactions Calculator. Nucleic acids Research 35: 473- 476.
- Ahmad S, Gromiha MM, Fawerah H and Sarai A (2004) ASAView: Database and tool for solvent accessibility representation in proteins. BMC Bioinformatics 5: 51.
- http://ailab.cs.iastate.edu/bfpred
- Swapna LS, Bhaskara RM, Sharma J, Srinivasan N (2012) Roles of residues in the interface of transient protein-protein complexes before complexation. Nature Scientific reports 2: 334.
- Ma B, Elkayam T, Wolfson H, Nussinov R (2003) Protein–protein interactions: Structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc. Natl. Acad. Sci. 100: 5772-5777.
- Kortemme T, Baker D (2002) A simple physical model for binding energy hot spots in protein–protein complexes. Proc. Natl. Acad. Sci. 99: 14116-14121.
- Verkhivker GM, Bouzida D, Gehlhaar DK, Rejto PA, Freer ST, et al. (2002) Monte Carlo simulations of the peptide recognition at the consensus binding site of the constant fragment of human immunoglobulin G: the energy landscape analysis of a hot spot at the intermolecular interface. Proteins 48: 539-557.
- Kozakov D, Hall DR, Chuang GY, Cencic R, Brenkea R, et al. (2011) Structural conservation of druggable hotspots in protein–protein interfaces. Proc. Natl. Acad. Sci. 108: 13528-13533.
Relevant Topics
- Algal Blooms
- Blue Carbon Sequestration
- Brackish Water
- Catfish
- Coral Bleaching
- Coral Reefs
- Deep Sea Fish
- Deep Sea Mining
- Ichthyoplankton
- Mangrove Ecosystem
- Marine Engineering
- Marine Fisheries
- Marine Mammal Research
- Marine Microbiome Analysis
- Marine Pollution
- Marine Reptiles
- Marine Science
- Ocean Currents
- Photoendosymbiosis
- Reef Biology
- Sea Food
- Sea Grass
- Sea Transportation
- Seaweed
Recommended Journals
Article Tools
Article Usage
- Total views: 3129
- [From(publication date):
October-2017 - Jul 15, 2025] - Breakdown by view type
- HTML page views : 2282
- PDF downloads : 847