Research Article Open Access
Prevalence of Potential Nosocomial Pathogens Isolated from Environments of Regional Hospital of Korca, Albania
Zhinzela Qyli*Department of Nursing, FSHNH, Fan S Noli University Korçë, Albania
- *Corresponding Author:
- Zhinzela Qyli
Department of Nursing
FSHNH, Fan S Noli University Korçë
Albania
Tel: 00355672613311
E-mail: zhinzelaqyli@gmail.com
Received date: April 19, 2017; Accepted date: May 09, 2017; Published date: May 16, 2017
Citation: Qyli Z (2017) Prevalence of Potential Nosocomial Pathogens Isolated from Environments of Regional Hospital of Korca, Albania. J Comm Pub Health Nurs 3:171. doi:10.4172/2471-9846.1000171
Copyright: © 2017 Qyli Z. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Visit for more related articles at Journal of Community & Public Health Nursing
Abstract
Background: Nosocomial infections make one of the most important issues regarding the surity of the patients at the healthcare institutions. Every hospitalized patient may acquire one nosocomial infection. The microorganisms that cause these infections may be bacteria, viruses and pathogen fungus. Nosocomial infections are the 5th causes of death in hospitals. A third of nosocomial infections are preventable.
Objective: The main purpose of this study was the identification of prevalence of potential nosocomial pathogens isolated from environments of Regional Hospital of Korca, Albania.
Material and methods: A total of 393 bacteria were isolated and identified from the hospital. The microbial identification was done with microscopy after Gram staining, colonies morphology and biochemistry.
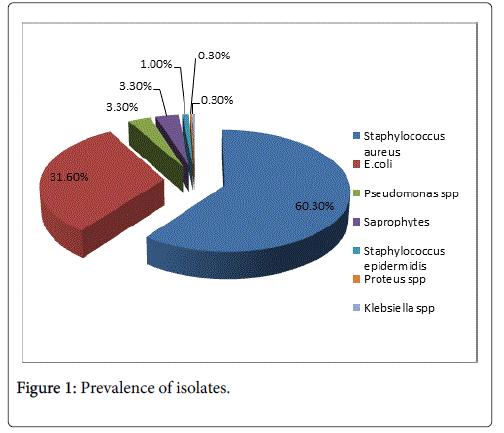
Results: The study revealed that the prevalence of microrganisms isolated was as following:
Staphylococcus aureus 237 (60.3%) of isolates, E. coli 124 (31.6%), Klebsiella spp. 1 (0.3%), Pseudomonas spp. 13 (3.3%), Proteus spp. 1 (0.3%), Staphylococcus epidermidis 4 (1.0%), Saprophytes 13 (3.3%).
Conclusion: Staphylococcus aureus was the most prevalent pathogen isolated in the hospitals while E. coli was the most frequent gram negative bacteria isolated.
Keywords
Nosocomial infection; hospital environment; Staphylococcus aureus; E. coli
Background
Nosocomial infections make one of the most important issues regarding the surity of the patients at the healthcare institutions. Every hospitalized patient may acquire one nosocomial infection. Hospitalacquired infections are a major challenge to patient safety. It is estimated that in 2002, a total of 1.7 million hospital-acquired infections occurred (4.5 per 100 admissions), and almost 99,000 deaths resulted from or were associated with a hospital-acquired infection [1], making hospital-acquired infections the sixth leading cause of death in the United States [2]; similar data have been reported from Europe [3]. The estimated costs to the U.S health care budget are $5 billion to $10 billion annually [4]. Approximately one third or more of hospitalacquired infections are preventable [5].
Hospital-acquired infections are the sixth leading cause of death in the United States and similar data have been reported from Europe [6], being an increasing problem in intensive care units (ICU), where the patients are more susceptible to colonization and the organisms are often more resistant to antimicrobial agents than in other environments. Since the 80's, infectious disease specialists have recognized that ICU patients acquire nosocomial infections at a much higher rate than patients elsewhere in the hospital [7]. In these confined areas, lower respiratory tract and bloodstream infections happen to be the most lethal; however, urinary tract infections are the most common. Infections caused by gram-negative bacteria have features that are of particular concern since these organisms are highly efficient at up-regulating or acquiring genes that code for mechanisms of antibiotic drug resistance, especially in the presence of antibiotic selection pressure. Most common nosocomial pathogens may well survive or persist on surfaces for a long time, what could explain their survival in ICU wards [8]. Gram-positive bacteria, such as Enterococcus spp. (including VRE), Staphylococcus aureus (including MRSA), Clostridium difficile or Streptococcus pyogenes, and many gram-negative bacteria, such as Acinetobacter spp., Klebsiella spp., Pseudomonas aeruginosa, Serratia marcescens or Shigella spp., may indeed survive for months on dry surfaces. A few others, such as Bordetella pertussis, Haemophilus influenzae , Proteus vulgaris or Vibrio cholerae , however, persist only for days [9,10].
Objective
The main purpose of this study was the identification of prevalence of potential nosocomial pathogens isolated from environments of Regional Hospital of Korca, Albania.
Materials and Methods
393 microbial isolates were obtained from various samples in the wards of the Regional Hospital of Korca. The items from which the samples were collected included sterilized materials, laundries, healthcare workers, surfaces, air and the systems of aspiration-intubation-oxygen. A swab soaked in nutrient broth was used to collect samples from the surfaces, laundries, health care workers and systems of aspiration-intubation-oxygen. All the samples were labeled properly and transported immediately to the microbiology laboratory of Korca for processing. The swab samples were inoculated in Blood agar, Sabouraud agar and incubated at 37°C for 24 h.
Air sampling was performed with settle plates methods. Petri dishes containing Blood and Sabouraud agar were placed at chosen places in the hospital environments at about 1 m above the ground and exposed for 15 min. After this exposure, the plates were covered with their lids and taken to laboratory in sealed plastic bags and incubated at 37°C for 24 h.
Identification of bacteria
After incubation bacterial isolates were identified according to the morphological appearance, cultural characteristics and biochemical reactions.
Bacterial colonies were viewed under a microscope after Gram staining. The procedure was as following: A slide of cell sample was made to be stained. Heat fixed the sample to the slide. Crystal violet was added to the slide and incubated for 1 min. The slide was rinsed with a gentle stream of water for a maximum of 5 s. Gram's iodine was added for 1 min, than the slide was rinsed again with water. The slide was rinsed with alcohol for ~3 s and with water. The sondary stain, safranin, was added to the slide and incubated for 1 min. The slide was rinsed with gentle stream of water for a maximum of 5 s.
Gram positive bacteria retained the prime stain (Crystal violet) and appeared violet under the microscope. Gram negative, lost the primary stain and took the sondary stain, causing it to appear red when viewed under a microscope.
Biochemical reactions were carried out in the isolates as follows
Catalase test
2-3 ml of hydrogen peroxide was purred in a test tube. A colony of test organism was taken with sterile wooden or glass rod and immersed it into hydrogen peroxide solution. Generation of bubbles indicated oxygen production. If bubbles were produced, the organism was catalase positive, if bubbles were not produced, the organism was catalase negative.
Oxidase test
A piece of filter paper was pleased in petri dish and 3 drops of freshly prepared oxidase reagent were added. Using a sterile glass rod, a colony of test organisms was removed from a culture plate and smeared on the filter paper.
Oxidase positive organisms gave blue color within 5-10 s, and in oxidase negative organisms, color did not change.
Citrate test
A bacterial colony was inoculated in simmons citrate agar and incubated at 35°C to 37°C for 18 to 24 h. Than development of blue color was observed.
Citrate positive
Growth was visible on the slant surface and the medium became an intense Prussian blue.
Citrate negative
Trace or no growth was visible and no color change occurred.
Indole test
Test bacterial colony were inoculated in tryptophan broth and incubated at 37°C for 24-28 h. Then 0.5 ml of Kovac’s reagent was added.
Positive test
Pink colored rink was observed after addition of reagent. Negative test: no color change after reagent addition.
Results
The study revealed that the prevalence of microorganisms isolated was as following: Staphylococcus aureus 237 (60.3%) of isolates, E. coli 124 (31.6%), Klebsiella spp. 1 (0.3%), Pseudomonas spp. 13 (3.3%), Proteus spp. 1 (0.3%), Staphylococcus epidermidis 4 (1.0%), Saprophytes 13 (3.3%) (Figure 1).
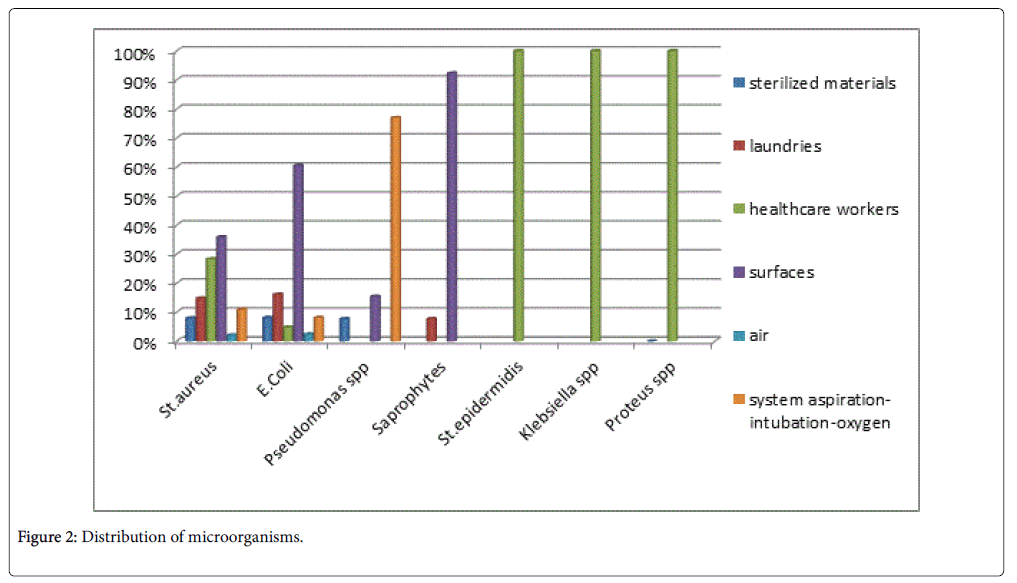
The distribution of Staphylococcus aureus isolates was as following:
8% of isolates were obtained from the sterilized materials, 14.8% from the laundries, 35.9% from the surfaces, 28.3% from healthcare workers, 2.1% from air and 11.0% from the systems of aspirationintubation- oxygen samples.
The distribution of E. coli isolates was as following:
8.1% of isolates were obtained from the sterilized materials, 16.1% from the laundries, 60.5% from the surfaces, 4.8% from healthcare workers, 2.4% from air and 8.1% from the systems of aspirationintubation- oxygen samples.
The distribution of Pseudomonas spp. isolates was as following:
7.7% of isolates were obtained from the sterilized materials, 15.4% from the surfaces and 76.9% from the systems of aspirationintubation- oxygen samples.
The distribution of Saprophytes isolates was as following:
7.7% from the laundries and 92.3% from the surfaces samples.
Staphylococcus epidermidis , Proteus spp. and Klebsiella spp. were isolated only from the healthcare workers samples (Table 1 and Figure 2).
| Sample | Â Microorganisms | |||||||
|---|---|---|---|---|---|---|---|---|
| S. aureus n (%) |
E. coli n (%) |
Klebsiellasp. n (%) | Pseudomonassp. n (%) | Proteussp. n (%) |
St.epidermidis n (%) | Saprophytes n (%) | Total n (%) | |
| Sterilized materials | 19 (8.0) | 10 (8.1) | 1 (7.7) | 30 (7.6) | ||||
| Laundries | 35 (14.8) | 20 (16.1) | 1 (7.7) | 56 (14.2) | ||||
| Healthcare workers | 67 (28.3) | 6 (4.8) | 1 (100.0) | 1 (100.0) | 4 (100.0) | 79 (20.1) | ||
| Surfaces | 85 (35.9) | 75 (60.5) | 2 (15.4) | 12 (92.3) | 174 (44.3) | |||
| Air | 5 (2.1) | 3 (2.4) | 8 (2.0) | |||||
| Systems of aspiration-intubation-oxygen | 26 (11.0) | 10 (8.1) | 10 (76.9) | 46 (11.7) | ||||
| Total n | 237 | 124 | 1 | 13 | 1 | 4 | 13 | 393 |
Table 1: Distribution of microorganisms.
Discussion
Identification of microorganisms of clinical importance in the environments of the Regional Hospital of Korca confirmed the presence of pathogens which are potentially related to nosocomial infections.
From the total of 393 isolates, the microorganism with the higher prevalence was Staphylococcus aureus 237 (60.3%) followed by E. coli 124 (31.6%), Klebsiella spp. 1 (0.3%), Pseudomonas spp. 13 (3.3%), Proteus spp. 1 (0.3%), Staphylococcus epidermidis 4 (1.0%), Saprophytes 13 (3.3%).
This result is similar to other studies that has reported S. aureus the most frequent bacteria in hospitals environments (60.30%) [11,12]. Staphylococcus aureus was isolated from all the types of samples, with a higher prevalence in the samples from the surfaces (35.9%).
E. coli was the most frequent gram negative bacteria isolated in hospital environments (31.60%) [13]. E. coli was isolated from all the types of samples, predominating in the samples from the surfaces (60.5%).
Pseudomonas spp. was the sond most frequent gram negative bacteria isolated in hospital environments (3.3%). Predomination of Pseudomonas spp. in the samples of the systems of aspirationintubation- oxygen (76.9%) indicates high risk for nosocomial pneumonia [14,15].
The prevalence of Saprophytes isolated from the hospital environments was 3.3%. They were mostly found in the samples from the surfaces (92.3%) [16-18].
Staphylococcus epidermidis, Klebsiella spp. and Proteus spp. were isolated with a low prevalence, respectively 1.0%, 0.3%, 0.3%. They were isolated only in the samples from the healthcare workers [19].
Conclusion
Hospital environment serve as a potencial reservoir of microorganisms that can cause nosocomial infections. The results of this study revealed that Staphylococcus aureus was the most prevalent pathogen isolated in the hospitals while E. coli was the most frequent gram negative bacteria isolated. The high prevalence of pathogens in this study might be due to poor hygienic practice in the hospital and lack of proper surveillance system for the control of potential nosocomial pathogens. Therefore, based on this finding it is recommended that management of the hospital should establish new politics which will help to minimize the spread of potential pathogens in the hospital environments.
References
- Klevens RM, Edwards JR, Richards CL Jr, Horan TC, Gaynes RP, et al. (2007) Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep 122: 160-166.
- Kung HC, Hoyert DL, Xu J, Murphy SL (2008) Deaths: Final data for 2005. Natl Vital Stat Rep 56: 1-120.
- Chopra I, Schofield C, Everett M, O'Neill A, Miller K, et al. (2008) Treatment of health-care-associated infections caused by gram-negative bacteria: A consensus statement. Lancet Infect Dis 8: 133-139
- Stone PW, Hedblom EC, Murphy DM, Miller SB (2005) The economic impact of infection control: Making the business case for increased infection control resources. Am J Infect Control 33: 542-547.
- Yokoe DS, Mermel LA, Anderson DJ, Arias KM, Burstin H, et al. (2008) A compendium of strategies to prevent healthcare-associated infections in acute care hospitals. Infect Control Hosp Epidemiol 1: S12-S21.
- Peleg AY, Hooper DC (2010) Hospital-acquired infections due to gram-negative bacteria. N Engl J Med 362: 1804-1813.
- Weber DJ, Raasch R, Rutala WA (1999) Nosocomial infections in the ICU: The growing importance of antibiotic-resistant pathogens. Chest 115: 34S-41S.
- Kramer A, Schwebke I, Kampf G (2006) How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis 6: 130.
- Weinstein RA, Nathan C, Gruensfelder R, Kabins SA (1980) Endemic aminoglycoside resistance in gram-negative bacilli: Epidemiology and mechanisms. J Infect Dis 141: 338-345.
- Maltezou HC, Drancourt M (2003) Nosocomial influenza in children. J Hosp Infect 55: 83-91.
- Okesola AO, Oni AA (2009) Antimicrobial resistance among common bacterial pathogens in south western Nigeria. American-Eurasian Journal of Agriculture and Environment Science 5: 327-330.
- Ijioma BC, Kalu IG and Nwachukwu CU (2010) Provide legend report and negative bacteria isolated in the hospital specimen. Rep Opin 2: 45-52.
- Cholley P, Thouverez M, Gbaguidi-Haore H, Sauget M, Slekovec C (2013) Hospital cross-transmission of extended-spectrum ÃÂ?-lactamase Escherichia coli and Klebsiella pneumoniae. Med Mal Infect 43: 331-336.
- Crnich CJ, Safdar N, Maki DG (2005) The role of the intensive care unit environment in the pathogenesis and prevention of ventilator-associated pneumonia. Respir Care 50: 813-836.
- Smith DL, Gumery LB, Smith EG, Stableforth DE, Kaufmann ME, et al. (1993) Epidemic of Pseudomonas cepacia in an adult cystic fibrosis unit: Evidence of person-to-person transmission. J Clin Microbiol 31: 3017-322.
- Konecka-Matyjek E, Mackiw E, Krygier B, Tomczuk K, Stos K, et al. (2012) National monitoring study of microbial contamination of food-contact surfacesin hospital kitchens in Poland. Ann Agric Environ Med 19: 457-463.
- Lajonchere JP, Feuilhade de Chauvin M (1994) Contamination by aspergillosis: Evaluation of preventive measures and monitoring of the environment. Pathol Biol (Paris) 42: 718-729.
- Faure O, Fricker-Hidalgo H, Lebeau B, Mallaret MR, Ambroise-Thomas P, et al. (2002) Eight-year surveillance of environmental fungal contamination in hospital operatin rooms and haematological units. J Hosp Infect 50: 155-160.
- Cholley P, Thouverez M, Gbaguidi-Haore H, Sauget M, Slekovec C, et al. (2013) Hospital cross-transmission of extended-spectrum ÃÂ?²-lactamase producing Escherichia coli and Klebsiella pneumoniae. Med Mal Infect 43: 331-336.
Relevant Topics
- Chronic Disease Management
- Community Based Nursing
- Community Health Assessment
- Community Health Nursing Care
- Community Nursing
- Community Nursing Care
- Community Nursing Diagnosis
- Community Nursing Intervention
- Core Functions Of Public Health Nursing
- Epidemiology
- Epidemiology in community nursing
- Health education
- Health Equity
- Health Promotion
- History Of Public Health Nursing
- Nursing Public Health
- Public Health Nursing
- Risk Factors And Burnout And Public Health Nursing
- Risk Factors and Burnout and Public Health Nursing
Recommended Journals
- Epidemiology journal
- Global Journal of Nursing & Forensic Studies
- Global Nursing & Forensic Studies Journal
- global journal of nursing & forensic studies
- journal of community medicine& health education
- journal of community medicine& health education
- Palliative Care & Medicine journal
- journal of pregnancy and child health
Article Tools
Article Usage
- Total views: 3316
- [From(publication date):
May-2017 - Dec 19, 2024] - Breakdown by view type
- HTML page views : 2578
- PDF downloads : 738