Research Article Open Access
Prevalence of Extended Spectrum Beta-lactamase Producing Enterobacteriaceae: A Cross Sectional Study at Adama Hospital, Adama, Ethiopia
GirmaMulisa1*, Lakew G Selassie1, Tilaye W1, Godana Jarso1, Teklu Shiferew1, Adinew Zewdu2, Wake Abebe2, Feleke Belachew2 and Tsegaye Sewunet31Adama Hospital Medical College, Adama, Ethiopia
2Oromia Public Health Regional Laboratory, Adama, Ethiopia
3Jimma University, Department of Laboratory Science and Pathology, Jimma Ethiopia
- *Corresponding Author:
- Girma Mulisa
Adama Hospital Medical College
Adama, Ethiopia
Tel: +251911928774; P.O. Box: 84
E-mail: girmamulisa30@gmail.com
Received date: October 09, 2015; Accepted date: November 28, 2015; Published date: December 14, 2015
Citation: Mulisa G, Selassie LG, Jarso G, Shiferew T, Zewdu A, et al. (2015) Prevalence of Extended Spectrum Beta-lactamase Producing Enterobacteriaceae: A Cross Sectional Study at Adama Hospital, Adama, Ethiopia. J Emerg Infect Dis 1:102. doi:10.4172/2472-4998.1000102
Copyright: © 2015 Mulisa G, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Infectious Disease and Pathology
Abstract
Background: The beta-lactam groups of antibiotics are the largest group of antibacterial agents used in clinical practice and they are frequently prescribed. The wide use of these antibiotics leads to the emergence and spread of resistant bacterial pathogens that produce the extended-spectrum β-lactamases (ESBL). Now days ESBLs producer bacteria are a common encounter in clinical practice and their prevalence varies from local to global. The aim of this study was to determine the prevalence of ESBL producing bacterial pathogens among Enterobacteriaceae isolates at Adama Hospital, Ethiopia.
Methods: Across sectional study was conducted from May 1, 2013-June 1, 2014 at Adama Teaching Hospital. A total of 384 consecutive non-repeat culture isolates were obtained from different clinical specimens of from patients visiting the Hospital. Bacterial strains were isolated following standard bacteriological techniques of American Society of microbiology. Antimicrobial susceptibility test was determined using Kirby-Bauer disk diffusion method and ESBL production was detected by modified double disc synergy test as stated on clinical and laboratory slandered institute. Data was analyzed using SPSS version 20.
Results: A total of 133 bacterial strains were isolated of which Enterobacteriaceae account for 68/133(51.1%). Twenty one isolates from the Enterobacteriaceae were suspected for ESBL production and 17/21 (80.95%) of them were confirmed to produce it with the overall prevalence of ESBL producers within the Enterobacteriaceae is 17/68 (25%). E. coli with the prevalence of 10/35 (28.57%) is the leading ESBL producer while Proteus species, Klebsiella species, E. cloacae and Citrobacter species accounted for 3/9 (33.3%), 2/8 (25%), 1/3 (33.3%) and 1/3 (33.3%) respectively.
Conclusion: The prevalence of ESBL producing Enterobacteriaceae was high among the clinical isolates of Adama Hospital Medical College. Because microbiology laboratories are limited in Ethiopia, the problem may go beyond expectation before it is unraveled. Thus routine screening of ESBL producing microorganisms from clinical samples should be considered where applicable.
Keywords
Extended-spectrum β-lactamases; Enterobacteriaceae; Antimicrobial agents
Abbreviations
CLSI: Clinical Laboratory Standard Institute; ESBL: Extended Spectrum Beta-lactamase; LIA: Lysine Iron Agar; SIM: Sulfide Indole Motility; TSI: Triple Sugar Iron; XLD: Xylose Lysine Deoxycholate
Introduction
Infectious diseases had been the leading cause of death until the middle of the 20th century [1] or until the introduction of novel antibiotics in clinical medicine which solved the problem of infectious diseases caused by bacterial pathogens [2]. The introduction of antibacterial agents was immediately accompanied by the emergence of drug resistant organisms which adapted different mechanisms to counter the effect of the antibiotics [3-7]. In the fight against the drug resistant strains of bacterial pathogens few potent drugs including the third generation cephalosporins have been discovered and have been in use for the treatment of various bacterial infections [8]. However the continual emergence of drug resistant bacterial pathogens including to the more potent third generation cephalosporin drugs made a great impediment to the rapid prognosis of patients in the hospital set up. This has been particularly of major clinical problem in the treatment of various infections caused by members of the Enterobacteriaceae family of bacterial pathogens.
The third generation cephalosporins antibiotics and the remaining β-lactam antibiotics are by far the largest group of antibacterial agents used in clinical medicine and they are among the most frequently prescribed antibiotics worldwide. They account for approximately 50% of global antibiotic consumption and with a few notable exceptions they remain the cornerstone of antibiotic therapy for a wide variety of systemic infections caused by bacterial pathogens [9]. However, the emergence and spread of extended-spectrum β-lactamases producing bacterial pathogens which are resistant to these potent antimicrobial agents including the third generation cephalosporins were compromised the use and wide applications of these novel antibiotics.
Antibiotic inactivation by the production of the specific enzyme which can break the most important functional group of the antibiotic namely the β-lactam ring is the most common mechanism of resistance that bacterial pathogens developed to β-lactam antibiotics. In Gram-negative bacterial pathogens, β-lactamase production is the major factor to resistance against the β-lactam antibiotics [10]. It inactivates the β-lactam antibiotics by hydrolysis of the β lactam ring resulting in ineffective compounds. One group of β-lactamases referred to extended-spectrum β-lactamases (ESBLs) has the ability to hydrolyze and cause resistance to various types of the newer β-lactam antibiotics, including the broad-spectrum cephalosporins [11]. Organisms that produce ESBLs remain an important reason for therapy failure with cephalosporins and have serious consequences for infection control [12]. In addition ESBL-producing organisms exhibit co-resistance to many other classes of non β lactam antibiotics [13] At the same time members of the Enterobacteriaceae are the most frequently encountered clinical isolates.
Magnitude of ESBL producer bacterial pathogen varies from country to country and even locally with in the same country. In Ethiopia there is a limitation of data on the prevalence of ESBLs producing bacterial pathogens. Hence, the study was aimed to determine the prevalence of β-lactamases producing bacterial pathogens within the family of Enterobacteriaceae.
Materials and Methods
Study area and setting
A laboratory based cross-sectional study design was used to conduct this study from September 01, 2013-May 30, 2014. Adama Hospital Medical College is found in Adama town at 95 km from Addis Ababa to the East. The hospital gives service for 800 patients per day at its outpatient department. The principal activity of the hospital is provision of health care but it is also engaged in medical education and research. The Hospital is serving a catchment population of more than 6 million; from regions (Oromia, Amhara, Afar, and Dire Dawa). The hospital has 212 beds capacity and serving on average 1000 per a day at six medical case teams (OPDs) and different specialty clinics. The hospital has 270 technical staff and 180 non-technical staffs.
Sample collection and antimicrobial susceptibility testing
Clinical samples were collected aseptically and processed at the microbiology laboratory of Oromia Regional Public Health Laboratory. The isolation and identification of bacterial isolates was conducted following standard bacteriological technique [14]. For isolation and identification of the following media were used: Blood agar, MacConky agar, XLD, TSI, LIA, SIM, Urea and Citrate. The bacterial isolates were then tested for antimicrobial susceptibility test by the disc diffusion method according to the guidelines of clinical and laboratory standard institute (CLSI) [15]. The following antibiotics discs were used in the sensitivity test; Cefotaxime (30 μg), Cefpodoxime (30 μg), Ceftrioxone (30 μg), Ceftazidime (30 μg), Cefepime (30 μg), Gentamicin(10 μg), Chloramphenicol (30 μg), Ciprofloxacin (5 μg), Nitrofurantoin (100 μg), Cotrimoxazole (25 μg), Piperacillin/Tazobactam (10 μg), and Tetracycline (10 μg) [Oxoid Uk].
Testing for the ESBL production
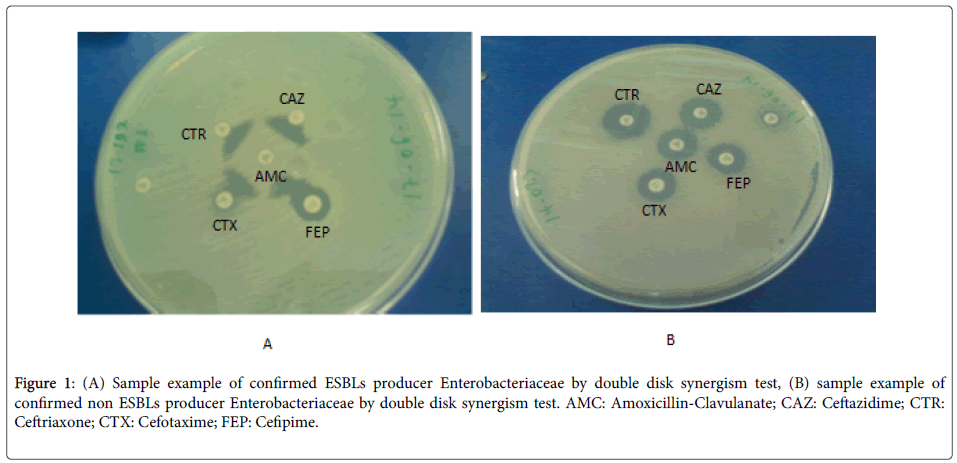
The ESBLs detection was carried out by modified double disc synergism test using Cefepime along with the third generation Cephalosporins [16]. All the strains which will show a diameter of less than 27 mm for Cefotaxime and less than 25 mm for Ceftriaxone were selected for checking the ESBLs production. The ESBL production was tested by the Modified Double Disc Synergy Test by using a disc of Amoxicillin-clavulanate (20/10 μg) along with four Cephalosporins (Cefotaxime, Ceftriaxone, Cefpodoxime and Cefepime). A lawn culture of the organisms was made on a Mueller-Hinton agar plate, as recommend by CLSI [17]. A disc which contained Amoxicillin-clavulanate (20/10 μg) was placed in the centre of the plate. The discs of third generation cephalosporin and fourth generation cephalosporin was placed 15 mm and 20 mm apart respectively, centre to centre to that of the Amoxicillin-clavulanatedisc [18]. Any distortion or increase in the zone towards the disc of Amoxicillin-clavulanatewas considered as positive for the ESBLs production (Figure 1).
Figure 1: (A) Sample example of confirmed ESBLs producer Enterobacteriaceae by double disk synergism test, (B) sample example of confirmed non ESBLs producer Enterobacteriaceae by double disk synergism test. AMC: Amoxicillin-Clavulanate; CAZ: Ceftazidime; CTR: Ceftriaxone; CTX: Cefotaxime; FEP: Cefipime.
Ethical consideration
Ethical clearance from e thical review board of Adama Hospital Medical College was obtained and the confidentiality of the patients was maintained. The results were communicated to the clinician or health care provider to initiate the recommended treatments based on susceptibility results.
Data analysis method
Data analysis was performed using SPSS ver. 20.0 and comparisons were made using binary logistic regression. Rates, types, and resistance pattern of the bacterial isolates with the respective antibiotics were calculated.
Results
A total of 384 samples were collected and processed for the isolation and identification of bacterial isolates. Of the total samples processed 133 were found to be culture positive and out of this Enterobacteriaceaeaccounted 68/133 (51.1%).
E. coli was the leading isolate of Enterobacteriaceae and followed by Proteus species. The processed clinical samples included urine, surgical wound swab, body fluids, corneal scrapping, nasal swab, stool and ear discharges. The majority of the isolates were from urine and surgical wound swab (Table 1).
| Specimen type | Organism | Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| E.coli | K. pneumoniae | Proteus species | Serraciae species | Citrobacter species | Morganella species | Entrococcus species | E. colaceae | Salmonella Species | ||
| Urine | 16 | 5 | 2 | 0 | 0 | 0 | 3 | 0 | 1 | 27 |
| Stool | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 3 |
| Surgical wound swab | 14 | 1 | 2 | 1 | 3 | 1 | 0 | 0 | 0 | 25 |
| Corneal scraping | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Pleural fluids | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| Ear discharge | 1 | 2 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 9 |
| Nasal swab | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Number (%) | 35(51.5) | 8(11.5) | 9(13.2) | 4(5.9) | 3(4.4) | 1(1.5) | 3(4.4) | 3(4.4) | 2(2.9) | 68(100) |
Table 1: Distribution of bacterial isolates from different clinical specimen.
Twenty three Enterobacteriaceae isolates were identified as potential ESBLs producers by screening method. Thus 17/68 (25%) were confirmed to ESBLs producer strains by phenotypic confirmatory test for ESBL production (Table 2).
| Isolated organisms | Number organism isolated | Confirmed ESBL produce N (%) |
|---|---|---|
| E.coli | 35 | 10(28.5) |
| K.pneumoniae | 8 | 2(25) |
| Proteus species | 9 | 3(33.3) |
| Citrobacter species | 3 | 1(33.3) |
| E. colaceae | 3 | 1(33.3) |
| Enterococcus species | 3 | 0 |
| S. salmonellae species | 2 | 0 |
| Morganellae species | 1 | 0 |
| Seraciae species | 4 | 0 |
| Total | 68 | 17(25) |
Table 2: Types of bacteria strains isolated as Enterobacteriaceae and their magnitude of ESBLs production.
E. coli and Proteus species were the leading agents of ESBL producers (Table 2).
These ESBLs producing strains were isolated from different clinical samples (urine, surgical wound swabs, body fluids and Ear discharges). Multiple ESBLs producing organisms were isolated from surgical wound swabs (Table 3).
| Organism | specimentype | Total | |||
|---|---|---|---|---|---|
| Urine | Wound swab | Pleural fluids | Ear discharge | ||
| E.coli | 2 | 5 | 3 | 0 | 10 |
| K. pneumonea | 0 | 1 | 0 | 1 | 2 |
| Proteus species | 2 | 1 | 0 | 0 | 3 |
| Citrobacter species | 0 | 1 | 0 | 0 | 1 |
| E. colaceae | 0 | 1 | 0 | 0 | 1 |
| Total | 4 | 9 | 3 | 1 | 17 |
Table 3: Distribution of ESBL producer bacterial isolates among clinical specimen.
The isolates of ESBLs producers were also found to be resistant to commonly used non β-lactam antibiotics including Gentamycin, Ciprofloxacin, Tetracycline, and Erythromycin (Table 4).
| ESBLs –phenotypic confirmatory testsorganisms(n) | |||||
|---|---|---|---|---|---|
| Antibiotics | E.coli (10) | K.pneumoniae(2) | Proteus species (3) | E. colaceae(1) | Citrobacter species (1) |
| TTC | 4 | 2 | |||
| SXT | 8 | 2 | 3 | 1 | |
| CXM | 5 | 1 | 1 | 1 | |
| CIP | 10 | 1 | 3 | ||
| GM | 7 | 1 | 2 | 1 | 1 |
Table 4: The multiple drug resistance patterns of the ESBLs producers within the family of the Enterobacteriaceae to the commonly used non β- lactam antibiotics. TTC: Tetracycline; SXT: Trimethoprim-sulfamethoxazole; CXM: Cefuroxime; CIP: Ciprofloxacin; GM: Gentamycin.
Discussion
The prevalence of ESBLs producers in this study was found to be 25%. The finding also indicated that the ESBLs producers were also resistant to multiple non β-lactam antibiotics including Gentamycin, Ciprofloxacin, Tetracycline and Erythromycin which are commonly used for the treatment of various kinds of infections. This should serve as a warning and precautionary notice for the physicians who are indiscriminately prescribing the third generation Cephalosporins and the other antibiotics based on empirical treatment for the management of various infections caused by pathogenic bacteria belonging to the family of Enterobacteriaceae.
A recent review on ESBLs producing pathogenic bacteria among the family Enterobacteriaceae in Africa indicated that the problem in health care institutions and communities is very large and revealed a significant increase in the prevalence in some countries in Africa [19]. In Ethiopia it was previously reported that 62.8% of the isolated Salmonella concord from stool and blood samples were ESBLs producers [20]. A study conducted at Jimma University Specialized Hospital indicated that the magnitude of ESBL producer E. coli and K. pneumoniae were 38.4% which is a higher prevalence compared to the finding in this study. It is anticipated due to the selected organisms were known for production of ESBL [21,22]. Studies conducted in Cameroon [23] and Brazil [24] reported the prevalence rate of 12% and 14.8% of ESBLs producers respectively among Enterobacteriaceae family. These prevalence rates are lower as compared to our study which may be attributed to variation in drug management policies and other local factors.
Proteus, E. coli and Klebsiella spp. were the leading ESBL producer in this study; other similar studies reported comparable results (similar isolates) [22,23,25]. Few numbers of Citrobacter and Entrobacter were also isolated that were found to be ESBLs producer strains. This finding showed that considerable number of the member of Enterobacteriaceae is potentially producing ESBLs. Thus ESBLs production within the family of Enterobacteriaceae may pose a serious problem in the management of the infections caused by pathogenic bacteria of this group. The most rational approach to the management of infections caused by these pathogenic bacteria is to rely on antibiotic sensitivity testing whenever the service is available or to use empirical treatment based on data available from studies that address the problem.
ESBL producers isolated in the study area were observed to be resistant to other class of antibiotics commonly used in addition to being resistant to the β Lactam drugs. Comparable results were reported before in Ethiopia and other countries [25,26]. Almost all isolated ESBL producers were resistant to Ciprofloxacin, Trimethoprim-sulfamethoxazole and Gentamycin. Similar findings were reported from other studies [27]. This may suggest that the production of ESBLs by pathogenic bacteria belonging to the family of Enterobacteriaceae may limit the treatment option available for the management of the infections caused by this group of bacteria. Thus the members of the family of Enterobacteriaceae that were proven to be produce ESBL pose a serious problem in the clinical management of the infections caused by them which trigger to put strong effort for the prevention ESBL occurrence.
Conclusion
As indicated in this study the prevalence of ESBLs producing bacteria among Enterobacteriaceae is high in the study area. The observed data demonstrated that this group of bacteria was also found out to be resistant to the non β-Lactam drugs which are commonly prescribed for the management of various infections. These dual properties of efficient mechanisms of drug resistance of being virtually unaffected by both the β-Lactam and the non β-Lactam antibiotics can confer a 100% protection for this group of bacteria. This problem may largely threaten developing countries like Ethiopia who are short of optional antimicrobials.
Hence the continual perpetuation of the pathogenic bacteria in causing illnesses and the menace of frustration of the physicians who find it very difficult in providing the much needed cure for the patient will continue as a vicious cycle. The vicious cycle could be disrupted when physicians will get the service for antibiotic sensitivity testing for the pathogenic bacteria they are trying to treat and they will prescribe the most effective antibiotic based on the antibiotic sensitivity profile presented for each pathogenic bacterial isolate.
Conflict of Interest
The authors declare that they have no competing interests.
Acknowledgement
We much appreciate Oromia Public Health Laboratory for provision of non-consumable materials that used for the laboratory isolation and characterization of the organisms and Adama Hospital Medical College for funding.
Authors’ Contributions
GM, conceived of the study, and participated in its design and coordination and drafts the manuscript. LG, GJ and TSh were participated in the design of the study and performed the statistical analysis. AZ and WA participated on sample collection, laboratory processing (isolation and identification of organism and phenotypic antibiotic susceptibility). FB and TS help to draft the manuscript and reviewed it. All authors read and approved the final manuscript.
References
- Bonner MS (2009) CA-MRSA the emerging.
- Rice LB (2008) Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis 197: 1079-1081
- Lyon RB, Skurray R (1987) Antimicrobial Resistance of Staphylococcus aureus: Genetic Basis. Microbiol Rev 51: 88-134
- Jevons PM (1961) To-day's drugs. British Medical Journal 14:1-2.
- Katayama Y, Ito T, Hiramatsu K (2000) A New Class of Genetic Element, Staphylococcus Cassette Chromosome mec, Encodes Methicillin Resistance in Staphylococcus aureus. Antimicrob Agents Chemother 44: 1549-1555.
- Panlilio AL, Culver DH, Gaynes RP, Banerjee S, Henderson TS, et al. (1992) Methicillin-resistant Staphylococcus aureus in U.S. hospitals. Infect Control HospEpidemiol 13:582-586.
- Turnidge J, Rao N, Chang YF, Fowler Jr VG, Kellie SM, et al. (2008) Staphylococcus aureus. Microbiology guided medline search.
- Grunden N. (2003) MRSA: a short history of a monster microbe. Pittsburgh Regional Healthcare Initiative:1-2.
- Brian I. Duerden (1987) Beta-Lactam Antibiotics in Systemic Infections. Phil J Microbiol Infect Dis 16:61-64.
- Livermore DM (2003) Bacterial resistance: origins, epidemiology, and impact. Clin Infect Dis 36: 11-23.
- Bradford PA (2001) Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. ClinMicrobiol Rev 14: 933-951.
- Paterson DL, Bonomo RA (2005) Extended-spectrum β-lactamases: a clinical update. ClinMicrobiol Rev 18: 657-686.
- Alipourfard I, Nili NY(2010) Antibiogram of Extended Spectrum Betalactamase (ESBL) producing Escherichia coli and Klebsiellapneumoniae isolated from Hospital Samples. Bangladesh J Med Microbiol 4: 32-36.
- Mindy P, Ajello G, Bopp C, Elliott J, Facklam R, et al. (2003) Manual for the Laboratory Identification and Antimicrobial Susceptibility Testing of Bacterial Pathogens of Public Health Importance in the Developing World. CDC and WHO: 1-383.
- Tzelepi E, Gaikkoupi P, SofianouD, Loukova V, Kemeroglou A, et al. (2000) J ClinMicrobiol 38:542-546.
- Kaur J, Chopra S, Sheevani, Mahajan G (2013) Modified Double Disc Synergy Test to Detect ESBL Production in Urinary Isolates of Escherichia coli and Klebsiella pneumonia. J ClinDiagn Res 7: 229-233.
- Wayne PA (2009) Performance standards for antimicrobial susceptibility testing; nineteenth informational supplement M100-S19. Clinical and Laboratory Standards Institute.
- Schmiedel J, Falgenhauer L, Domann E, Bauerfeind R, Prenger-Berninghoff E, et al. (2014) Multiresistant extended-spectrum β-lactamaseproducingEnterobacteriaceae from humans, companion animals and horses in central Hesse, Germany. BMC Microbiology 14:1-13.
- Livermore DM (1995) β-Lactamases in laboratory and clinical resistance. ClinMicrobiol Rev 8: 557-584.
- Moga Sh. Ali S, Wodafrash B (2014) Extended Expectrum Beta lactamase Production and antimicrobial Resistance in K. pneumoniae and E.coli among inpatients out patient of Jimma University specialized Hospital, South West Ethiopia. Afr. J. Microbiol. Res 8: 3687-3694.
- Storberg V (2014) ESBL-producing Enterobacteriaceae in Africa a non-systematic literature review of research published 2008-2012. Infect EcolEpidemiol 4: 1-16.
- Beyene G, Nair S, AsratD, Mengistu Y, Engers H, et al. (2011) Multidrug resistant Salmonella concord is a major cause of salmonellosis in children in Ethiopia. J Infect DevCtries 5: 23-33.
- Kandeel A (2014) Prevalence and risk factors of extended-spectrum β-lactamases producing Enterobacteriaceae in a general hospital in Saudi Arabia. JMID 4: 50-54.
- Gangoué-Piéboji J, Bedenic B, Koulla-Shiro S, Randegger C, Adiogo D, et al. (2005) Extended-Spectrum Lactamase-Producing Enterobacteriaceae in Yaounde, Cameroon. J ClinMicrobiol 47: 3273-3277.
- Abreu AG, Marques SG, Monteiro-Neto V, Gonçalves AG(2013) Extended-spectrum -lactamase-producing enterobacteriaceae in community-acquired urinary tract infections in São Luís, Brazil. Braz J Microbiol 44: 469-471.
- Rakotonirina HC, Garin B, Randrianirina F, Richard V, Talarmin A, et al. (2013) Molecular characterization of multidrug-resistant extended-spectrum β-lactamase-producing Enterobacteriaceae isolated in Antananarivo, Madagascar. BMC Microbiology 13:1-10.
- Deodurg PM, Sureka RK, Mala RD (2014) Prevalence and antibiogram of extended spectrum β- lactamase producing Klebsiellapneumoniaein a tertiary care hospital. Journal of Scientific and Innovative Research 3: 155-159.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 14093
- [From(publication date):
March-2016 - Apr 02, 2025] - Breakdown by view type
- HTML page views : 13031
- PDF downloads : 1062