Research Article Open Access
Preliminary Set Theory-Type Analysis of Proteins Associated With Parkinson's Disease
Paul Whitesman*
Disease Motifs, Rochdale, Greater Manchester, England
- Corresponding Author:
- Paul Whitesman
Disease Motifs, Rochdale, Greater Manchester, England
Tel: ++44 (0) 1706 343120
E-mail: paulwhitesman@diseasemotifs.co.uk
Received date: September 29, 2014; Accepted date: November 11, 2014; Published date: November 18, 2014
Citation: Whitesman P (2014) Preliminary Set Theory-Type Analysis of Proteins Associated With Parkinson’s Disease. J Alzheimers Dis Parkinsonism 4:170. doi: 10.4172/2161-0460.1000170
Copyright: © 2014 Whitesman P. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Alzheimers Disease & Parkinsonism
Abstract
In an attempt to create a model of Parkinson’s disease (PD) eighty-three proteins were extracted from the Swiss- Prot protein database that had some casual mention of PD. These were split up into various subsets of proteins of which three are focused on here: PARK, made up of proteins that had some indication that polymorphisms in the protein might increase a person’s susceptibility to develop PD; MITOCHOND, proteins which had some association with the mitochondria; and MT-C1D, proteins that were implicated in mitochondrial complex 1 deficiency. The PARK subset had 21 out of 83 proteins (21/83); MITOCHOND 33 out of 83 proteins (33/83); and MT-C1D 17 out of 83 proteins (17/83). The results could be used to build up a basic model of PD creating phenotypes based on sets of proteins. The main phenotypes established here are; non-mitochondrial PD (50/83) and mitochondrial PD (33/83). Further division is possible dependant on whether proteins have polymorphisms which increase susceptibility to develop PD. MT-C1D seems to be independent of the PARK set. This is a very simplistic attempt at trying to model Parkinson’s disease at the proteomic level and will need further work to build up the more complex and realistic PD proteomic disease model.
Keywords
Parkinson’s disease- PD, Mitochondrial complex 1 deficiency- MT-C1D, Mitochondria set theory-type analysis, Proteomic set theory-type analysis, Disease networks, Disease networking, Networks, Disease motifs
Introduction
Various diseases are known to have some genetic polymorphism that increases the likelihood or susceptibility of developing a disease. There are some proteins which are in some way linked to a particular disease by either: a polymorphism that leads to an increased susceptibility; increasingly or decreasingly expressed in a particular disease; or the protein is shown in some pathological symptom, as in the case of Parkinson’s disease (PD) where alpha-synuclein can form parts of Lewy bodies [1].
This is an introduction to ‘set theory-type’ analysis of some proteins associated with Parkinson’s disease. The set theory-type proteomic analysis here is less about a mathematical treatment of the elucidation of the pathological process of PD but only a simplistic attempt at grouping proteins into sets so as to get a clear picture of the possible pathology involved in the disease process. This disease process can be referred to as a disease network of interacting proteins. It also may be used to form some proteomic foundation for categorising particular phenotypes attributed to a particular disease type.
This work focuses on PD and in which there is evidence that there is some involvement with the mitochondria [2]. There are also diseases such as mitochondrial complex 1 deficiency MT-C1D which, as well as been a condition in its own right, has been associated with progression to PD [3-5].
Method
The Swiss-Prot human protein database was downloaded from the EBI website on 29th October 2014. A Perl script was written that extracted any protein data-sheet that had mention of ‘Parkinson’ in the comments section (CC) or feature table (FT) of the data-sheet. Originally 86 proteins were found but three of these, AAKG2_ HUMAN, FGF20_HUMAN and SYUB_HUMAN, were considered to be false positives.
Most of the 83 proteins did not have any known polymorphisms that would lead to an increased susceptibility to develop PD but just mentioned that in some way they were implicated in the disease. From this initial set of 83 proteins which were deemed to have some association with PD a series of over sixty different subsets were produced. Some of these may well be labelled pseudo-sets as these were based on terms that may have been written into the Swiss-Prot datasheet entry in a superficial manner such as mentioning all the terms associated with PD such as ataxia, fatigue, dystonia etc. when in fact each particular term may not have any real relevance to that actually specific protein but is just a generalised description of PD.
The three subsets, which all derived from the original 83 PD proteins, are: MITOCHOND, which consists of the mitochondrial proteins; PARK, these were the proteins that had at least one actual polymorphism that is believed might lead to an increased susceptibility to developing PD; and MT-C1D, which included proteins implicated in Mitochondrial complex 1 deficiency. MITOCHOND and MT-C1D were extracted by use of a Perl script, as mentioned above, looking in the CC and FT sections while the PARK was extracted by just looking at the FT section, at the variants. These subsets were compared and contrasted by use of another Perl script to form ‘AND’ sets e.g. PARK AND MITOCHOND.
For further details of these sets and for the free use of the Perl scripts (available on request) please visit Disease Motifs at http://www.diseasemotifs.co.uk
Results
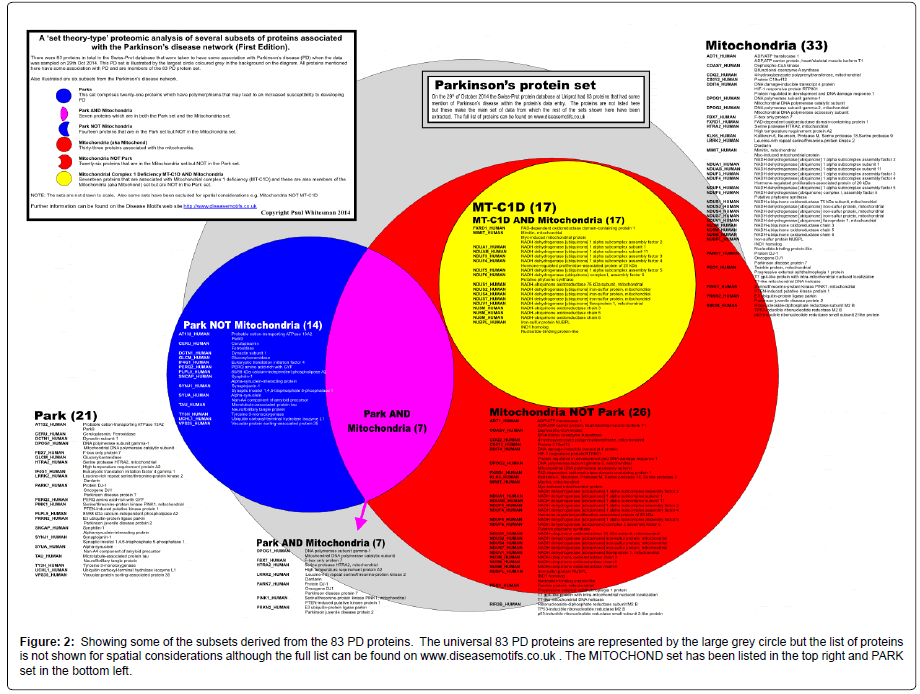
Out of the 83 proteins that were initially established as the PD set; MITOCHOND had 33 proteins (33/83); PARK had 21 proteins (21/83); and MT-C1D had 17 proteins (17/83).
Seven of the PARK set were in the MITOCHOND (mitochondrial) set (7/83) and hence fourteen proteins from the PARK set were NOT in the MITOCHOND set i.e. not mitochondrial proteins, PARK NOT MITOCHOND (14/83). The seventeen proteins that are in the MTC1D set were totally included in the MITOCHOND set while none of the MT-C1D proteins were found in the PARK set.
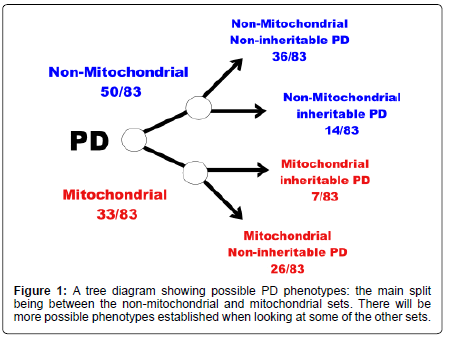
The conclusion of this is that perhaps one can split the proteins associated with PD into at least two groups depending on whether they have some association with the mitochondria or not. Further division is possible based on whether the proteins have been shown to have some polymorphisms leading to an increased susceptibility to develop PD (Figures 1 and 2). MT-C1D is independent of proteins that have been shown to increase susceptibility to develop PD (Table 1-6).
Discussion
The purpose of this work is to try and look at the proteins associated with PD and to try gained some understanding of the nature of PD by looking at these proteins. It has not been the purpose of this work to look in detail at the actual proteins sequences themselves only some secondary characteristics have been used to group these proteins into sets. There have been over sixty of such sets that have been created (see ‘Set Theory’ page in PD section at www.diseasemotifs.co.uk).
Figure 2: Showing some of the subsets derived from the 83 PD proteins. The universal 83 PD proteins are represented by the large grey circle but the list of proteins is not shown for spatial considerations although the full list can be found on www.diseasemotifs.co.uk . The MITOCHOND set has been listed in the top right and PARK set in the bottom left.
| 1 | AT132_HUMAN | Probable cation-transporting ATPase 13A2; Park9 |
| 2 | CERU_HUMAN | Ceruloplasmin; Ferroxidase |
| 3 | DCTN1_HUMAN | Dynactin subunit 1 |
| 4 | DPOG1_HUMAN | DNA polymerase subunit gamma-1; Mitochondrial DNA polymerase catalytic sub unit |
| 5 | FBX7_HUMAN | F-box only protein 7 |
| 6 | GLCM_HUMAN | Glucosylceramidase |
| 7 | HTRA2_HUMAN | Serine protease HTRA2, mitochondrial; High temperature requirement protein A2 |
| 8 | IF4G1_HUMAN | Eukaryotic translation initiation factor 4 gamma 1 |
| 9 | LRRK2_HUMAN | Leucine-rich repeat serine/threonine-protein kinase 2; Dardarin |
| 10 | PARK7_HUMAN | Protein DJ-1; Oncogene DJ1; Parkinson disease protein 7 |
| 11 | PERQ2_HUMAN | PERQ amino acid-rich with GYF |
| 12 | PINK1_HUMAN | Serine/threonine-protein kinase PINK1; mitochondrial; PTEN-induced putative kinase protein 1 |
| 13 | PLPL9_HUMAN | 85/88 kDa calcium-independent phospholipase A2 |
| 14 | PRKN2_HUMAN | E3 ubiquitin-protein ligase parkin; Parkinson juvenile disease protein 2 |
| 15 | SNCAP_HUMAN | Synphilin-1; Alpha-synuclein-interacting protein |
| 16 | SYNJ1_HUMAN | Synaptojanin-1 |
| Synaptic inositol 1,4,5-trisphosphate 5-phosphatase 1 | ||
| 17 | SYUA_HUMAN | Alpha-synuclein; Non-A4 component of amyloid precursor |
| 18 | TAU_HUMAN | Microtubule-associated protein tau; Neurofibrillary tangle protein |
| 19 | TY3H_HUMAN | Tyrosine 3-monooxygenase |
| 20 | UCHL1_HUMAN | Ubiquitin carboxyl-terminal hydrolase isozyme L1 |
| 21 | VPS35_HUMAN | Vacuolar protein sorting-associated protein 35 |
Table 1: PARK – 21 proteins that have some indication that some polymorphisms may lead to an increased susceptibility to develop PD. Swiss-Prot protein ID and descriptive names shown.
| 1 | DPOG1_HUMAN | DNA polymerase subunit gamma-1; Mitochondrial DNA polymerase catalytic subunit |
| 2 | FBX7_HUMAN | F-box only protein 7 |
| 3 | HTRA2_HUMAN | Serine protease HTRA2, mitochondrial; High temperature requirement protein A2 |
| 4 | LRRK2_HUMAN | Leucine-rich repeat serine/threonine-protein kinase 2; Dardarin |
| 5 | PARK7_HUMAN | Protein DJ-1; Oncogene DJ1; Parkinson disease protein 7 |
| 6 | PINK1_HUMAN | Serine/threonine-protein kinase PINK1, mitochondrial; PTEN-induced putative kinase protein 1 |
| 7 | PRKN2_HUMAN | E3 ubiquitin-protein ligase parkin; Parkinson juvenile disease protein 2 |
Table 2: PARK AND MITOCHOND – 7 proteins that have some indication that some polymorphisms may lead to an increased susceptibility to develop PD AND have some connection to the mitochondria. Swiss-Prot protein ID and descriptive names shown.
The main consideration here is the connection between PD and the mitochondria and to perhaps differentiate between two, three or more types of PD based on this grouping method. Is it a realistic proposition to group mitochondrial proteins together and label this as a mitochondrial form of PD and can this be matched in clinic considerations?
One of the main problems with the work is the naïve assumption that you can build up a set identity by using the, sometimes subjective, literature that is contained within the comments (CC) of the data-sheets that are in the Swiss-Prot protein database. Fixing on a search term such as ‘fatigue’ which may be used as a term superficially to describe the symptoms of PD in general when commenting in the Swiss-Prot datasheet, rather than as a term that has actually any real relevance to that particular protein. This unreal attachment of this term to a group of proteins creates the idea of a pseudo-set i.e. a group of proteins that have a superficial term as their grouping factor. Even some of the 83 proteins in the main PD set may be considered to be superficial linked to PD.
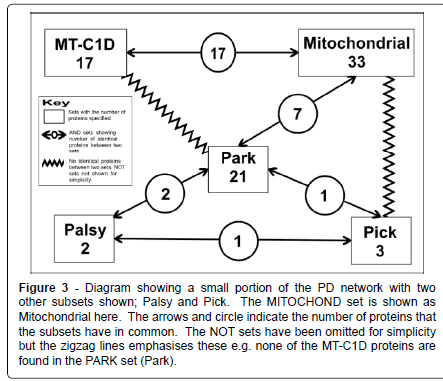
Figure 3: Diagram showing a small portion of the PD network with two other subsets shown; Palsy and Pick. The MITOCHOND set is shown as Mitochondrial here. The arrows and circle indicate the number of proteins that the subsets have in common. The NOT sets have been omitted for simplicity but the zigzag lines emphasises these e.g. none of the MT-C1D proteins are found in the PARK set (Park).
However there is surely some benefit on establishing whether a protein: is a mitochondrial protein or not; whether a group of proteins can be placed in one set due to their involvement in some process of a particular disease such as PD, MT-C1D, cancer or some other disease; or whether there is some similarity of function among a group of proteins e.g. ferroxidase. All the proteins looked at here have some association with PD. Here we are looking mainly at three subsets taken from this larger set of 83 PD proteins: PARK, proteins that are known to have some genetic polymorphisms that will lead to an actual increased susceptibility to develop PD; MITOCHOND made of proteins that had some connection with the mitochondria; and MT-C1D made up of proteins that are associated with mitochondrial complex 1 deficiency (Figure 3).
| 1. | AT132_HUMAN | Probable cation-transporting ATPase 13A2; Park9 |
| 2. | CERU_HUMAN | Ceruloplasmin; Ferroxidase |
| 3. | DCTN1_HUMAN | Dynactin subunit 1 |
| 4. | GLCM_HUMAN | Glucosylceramidase |
| 5. | IF4G1_HUMAN | Eukaryotic translation initiation factor 4 |
| 6. | PERQ2_HUMAN | PERQ amino acid-rich with GYF |
| 7. | PLPL9_HUMAN | 85/88 kDa calcium-independent phospholipase A2 |
| 8. | SNCAP_HUMAN | Synphilin-1; Alpha-synuclein-interacting protein |
| 9. | SYNJ1_HUMAN | Synaptojanin-1 |
| Synaptic inositol 1,4,5-trisphosphate 5-phosphatase 1 | ||
| 10. | SYUA_HUMAN | Alpha-synuclein; Non-A4 component of amyloid precursor |
| 11. | TAU_HUMAN | Microtubule-associated protein tau; Neurofibrillary tangle protein |
| 12. | TY3H_HUMAN | Tyrosine 3-monooxygenase |
| 13. | UCHL1_HUMAN | Ubiquitin carboxyl-terminal hydrolase isozyme L1 |
| 14. | VPS35_HUMAN | Vacuolar protein sorting-associated protein 35 |
Table 3: PARK NOT MITOCHOND 14 proteins that have some indication that some polymorphisms may lead to an increased susceptibility to develop PD but which are not obvious mitochondrial proteins. Swiss-Prot protein ID and descriptive names shown.
| 1. | ADT1_HUMAN | ADP/ATP translocase 1; ADP,ATP carrier protein, heart/skeletal muscle isoform T1 |
| 2. | COASY_HUMAN | Dephospho-CoA kinase |
| Bifunctional coenzyme A synthase | ||
| 3. | COQ2_HUMAN | 4-hydroxybenzoate polyprenyltransferase, mitochondrial |
| 4. | CS012_HUMAN | Protein C19orf12 |
| 5. | DDIT4_HUMAN | DNA damage-inducible transcript 4 protein; HIF-1 responsive protein RTP801; |
| Protein regulated indevelopment and DNA damage response 1 | ||
| 6. | DPOG1_HUMAN | DNA polymerase subunit gamma-1; Mitochondrial DNA polymerase catalytic subunit |
| 7. | DPOG2_HUMAN | DNA polymerase subunit gamma-2, mitochondrial; |
| Mitochondrial DNA polymerase accessory subunit | ||
| 8. | FBX7_HUMAN | F-box only protein 7 |
| 9. | FXRD1_HUMAN | FAD-dependent oxidoreductase domain-containing protein 1 |
| 10. | HTRA2_HUMAN | Serine protease HTRA2, mitochondrial; High temperature requirement protein A2 |
| 11. | KLK6_HUMAN | Kallikrein-6; Neurosin; Protease M; Serine protease 18; Serine protease 9 |
| 12. | LRRK2_HUMAN | Leucine-rich repeat serine/threonine-protein kinase 2; Dardarin |
| 13. | MIMIT_HUMAN | Mimitin, Myc-induced mitochondrial protein; |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 2 | ||
| 14. | NDUA1_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 1 |
| 15. | NDUAB_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 11 |
| 16. | NDUF3_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3 |
| 17. | NDUF4_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 4 |
| Hormone-regulated proliferation-associated protein of 20 kDa | ||
| 18. | NDUF5_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 5 |
| 19. | NDUF6_HUMAN | NADH dehydrogenase (ubiquinone) complex I, assembly factor 6; Putative phytoene synthase |
| 20. | NDUS1_HUMAN | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial |
| 21. | NDUS2_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein, mitochondrial |
| 22. | NDUS4_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein, mitochondrial |
| 23. | NDUS7_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein, mitochondrial |
| 24. | NDUV1_HUMAN | NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial |
| 25. | NU3M_HUMAN | NADH-ubiquinone oxidoreductase chain 3 |
| 26. | NU5M_HUMAN | NADH-ubiquinone oxidoreductase chain 5 |
| 27. | NU6M_HUMAN | NADH-ubiquinone oxidoreductase chain 6 |
| 28. | NUBPL_HUMAN | Iron-sulfur protein NUBPL; IND1 homolog; Nucleotide-binding protein-like |
| 29. | PARK7_HUMAN | Protein DJ-1; Oncogene DJ1; Parkinson disease protein 7 |
| 30. | PEO1_HUMAN | Twinkle protein, mitochondrial; Progressive external ophthalmoplegia 1 protein; |
| T7 gp4-like protein with intra-mitochondrial nucleoid localization; | ||
| T7-like mitochondrial DNA helicase | ||
| 31. | PINK1_HUMAN | Serine/threonine-protein kinase PINK1; PTEN-induced putative kinase protein 1 |
| 32. | PRKN2_HUMAN | E3 ubiquitin-protein ligase parkin; Parkinson juvenile disease protein 2 |
| 33. | RIR2B_HUMAN | Ribonucleoside-diphosphate reductase subunit M2 B; |
| TP53-inducible ribonucleotide reductase M2 B; | ||
| p53-inducible ribonucleotide reductase small subunit 2-like protein |
Table 4: MITOCHOND [aka MITOCHONDRIAL] – 33 proteins that have some connection to the mitochondria. Swiss-Prot protein ID and descriptive names shown.
| 1. | FXRD1_HUMAN | FAD-dependent oxidoreductase domain-containing protein 1 |
| 2. | MIMIT_HUMAN | Mimitin; Myc-induced mitochondrial protein |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 2 | ||
| 3. | NDUA1_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 1 |
| 4. | NDUAB_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 11 |
| 5. | NDUF3_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3 |
| 6. | NDUF4_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 4 |
| Hormone-regulated proliferation-associated protein of 20 kDa | ||
| 7. | NDUF5_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 5 |
| 8. | NDUF6_HUMAN | NADH dehydrogenase (ubiquinone) complex I, assembly factor 6; Putative phytoene synthase |
| 9. | NDUS1_HUMAN | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial |
| 10. | NDUS2_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein, mitochondrial |
| 11. | NDUS4_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein, mitochondrial |
| 12. | NDUS7_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein, mitochondrial |
| 13. | NDUV1_HUMAN | NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial |
| 14. | NU3M_HUMAN | NADH-ubiquinone oxidoreductase chain 3 |
| 15. | NU5M_HUMAN | NADH-ubiquinone oxidoreductase chain 5 |
| 16. | NU6M_HUMAN | NADH-ubiquinone oxidoreductase chain 6 |
| 17. | NUBPL_HUMAN | Iron-sulfur protein NUBPL; IND1 homolog; Nucleotide-binding protein-like |
Table 5: MT–C1D 17 proteins that have some indication that some polymorphisms may lead to an increased susceptibility to develop Mitochondrial complex 1 deficiency (MT-C1D). Swiss-Prot protein ID and descriptive names shown.
One observation is that out of the 83 proteins in the entire collection of PD proteins just less than a half of these have some connection with the mitochondria which might suggest the importance of the role of mitochondrial proteins in PD. On the other hand though the greater part of the PARK group, which is the group of proteins that contain proteins that have some polymorphisms associated with an increase in susceptibility to develop PD, have no obvious connection with the mitochondria.
From this part of the disease network it might be possible to infer that perhaps we could divide PD into at least two major components: non-mitochondrial PD; and mitochondrial PD. The non-mitochondrial PD would include the PARK NOT MITOCHOND subset, based on the 14 proteins in that set, this is with proteins that have no obvious connection with the mitochondria. The second group, mitochondrial PD, could be split into two or more further groups: one that has the genetic susceptibility to develop PD involving the seven proteins in the PARK AND MITOCHOND subset; the other is the residual proteins in the MITOCHOND NOT PARK subset which includes the MTC1D subset which does not contain proteins that have been shown to have some capacity to alter the susceptibility to develop PD but may increase susceptibility in the development of another disease state (e.g. MT-C1D). Presumably there are other sets outside of the PARK and MITOCHOND sets that can lead to the development of PD: which have no inheritable PD tendency nor have any connection with the mitochondria. MT-C1D does not include any proteins from the PARK set which may suggest that when PD develops from the MT-C1D pathway this could be characterised as a non-inheritable PD phenotype.
| 1. | ADT1_HUMAN | ADP/ATP translocase 1; ADP,ATP carrier protein, heart/skeletal muscle isoform T1 |
| 2. | COASY_HUMAN | Dephospho-CoA kinase |
| Bifunctional coenzyme A synthase | ||
| 3. | COQ2_HUMAN | 4-hydroxybenzoate polyprenyltransferase, mitochondrial |
| 4. | CS012_HUMAN | Protein C19orf12 |
| 5. | DDIT4_HUMAN | DNA damage-inducible transcript 4 protein; HIF-1 responsive protein RTP801; |
| Protein regulated in development and DNA damage response 1 | ||
| 6. | DPOG2_HUMAN | DNA polymerase subunit gamma-2, mitochondrial; |
| Mitochondrial DNA polymerase accessory subunit | ||
| 7. | FXRD1_HUMAN | FAD-dependent oxidoreductase domain-containing protein 1 |
| 8. | KLK6_HUMAN | Kallikrein-6; Neurosin; Protease M; Serine protease 18; Serine protease 9 |
| 9. | MIMIT_HUMAN | Mimitin; Myc-induced mitochondrial protein; |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 2 | ||
| 10. | NDUA1_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 1 |
| 11. | NDUAB_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 11 |
| 12. | NDUF3_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3 |
| 13. | NDUF4_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 4 |
| Hormone-regulated proliferation-associated protein of 20 kDa | ||
| 14. | NDUF5_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 5 |
| 15. | NDUF6_HUMAN | NADH dehydrogenase (ubiquinone) complex I, assembly factor 6; Putative phytoene synthase |
| 16. | NDUS1_HUMAN | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial |
| 17. | NDUS2_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein, mitochondrial |
| 18. | NDUS4_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein, mitochondrial |
| 19. | NDUS7_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein, mitochondrial |
| 20. | NDUV1_HUMAN | NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial |
| 21. | NU3M_HUMAN | NADH-ubiquinone oxidoreductase chain 3 |
| 22. | NU5M_HUMAN | NADH-ubiquinone oxidoreductase chain 5 |
| 23. | NU6M_HUMAN | NADH-ubiquinone oxidoreductase chain 6 |
| 24. | NUBPL_HUMAN | Iron-sulfur protein NUBPL; IND1 homolog; Nucleotide-binding protein-like |
| 25. | PEO1_HUMAN | Twinkle protein; Progressive external ophthalmoplegia 1 protein; |
| T7 gp4-like protein with intra-mitochondrial nucleoid localization; | ||
| T7-like mitochondrial DNA helicase | ||
| 26. | RIR2B_HUMAN | Ribonucleoside-diphosphate reductase subunit M2 B |
| TP53-inducible ribonucleotide reductase M2 B | ||
| p53-inducible ribonucleotide reductase small subunit 2-like protein |
Table 6: MITOCHOND NOT PARK 26 proteins that have some connection with the mitochondria but are not in the PARK set. Swiss-Prot protein ID and descriptive names shown.
It is likely that there are additions (and possibly subtractions from errors made in this study) to be made to the number and type of proteins involved in each set. The actual numbers in each set does not really matter and neither do some of the errors as it is the conclusions from the consensus of each set that is important. One major error is the missing of alpha-synuclein as being associated with the mitochondria as it has been shown to be localised to mitochondria [6]. Maybe using the GO terms would be more effective, at least for some terms, and some attempts have made to do this.
Summary and conclusion
It may be possible to split PD into categories or types based on particular protein characteristic:
I. Mitochondrial inheritable (increased susceptibility) PD (7/83);
II. Mitochondrial non-inheritable PD (26/83);
III. Non-mitochondrial inheritable PD (14/83);
IV. And non-mitochondrial non-inheritable PD (36/83).
Whether this can be translated into something that is clinically useful is debatable. Perhaps one of the symptoms of PD such as chronic ‘fatigue’ could suggest that the person with this ‘type’ of PD has a form of mitochondrial PD. If this were the case then one could perhaps tailor PD medication to suit the particular type of PD.
References
- Baba M, Nakajo S, Tu PH, Tomita T, Nakaya K, et al. (1998) Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson's disease and dementia with Lewy bodies. Am J Pathol 152: 879-884.
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, et al. (2004) Hereditary early-onset Parkinson's disease caused by mutations in PINK1. 304: 1158-1160.
- Schapira A.H et al. (1989) Mitochondrial Complex I Deficiency In Parkinson's Disease. Lancet 1: 1269
- Schapira A.H (1990) et al. Mitochondrial complex I deficiency in Parkinson's disease. J Neurochem, 54: 823-827
- Schapira AH (1993) Mitochondrial complex I deficiency in Parkinson's disease. Adv Neurol 60: 288-291
- Li WW, Yang R, Guo JC, Ren HM, Zha XL, et al. (2007) Localization of alpha-synuclein to mitochondria within midbrain of mice. Neuroreport 18: 1543-1546
Relevant Topics
- Advanced Parkinson Treatment
- Advances in Alzheimers Therapy
- Alzheimers Medicine
- Alzheimers Products & Market Analysis
- Alzheimers Symptoms
- Degenerative Disorders
- Diagnostic Alzheimer
- Parkinson
- Parkinsonism Diagnosis
- Parkinsonism Gene Therapy
- Parkinsonism Stages and Treatment
- Stem cell Treatment Parkinson
Recommended Journals
Article Tools
Article Usage
- Total views: 15791
- [From(publication date):
December-2014 - Jul 11, 2025] - Breakdown by view type
- HTML page views : 11123
- PDF downloads : 4668