Potential for Bio Control of Fungal Phytopathogens by Pseudomonas Aeruginosa CFSP1L1 Isolated from a Matured Compost Sample
Received: 09-Aug-2021 / Accepted Date: 23-Aug-2021 / Published Date: 30-Aug-2021 DOI: 10.4172/2155-6199.1000004
Abstract
The use of antimicrobial bioactive compounds is currently regarded as a novel technology for biological control against plant diseases. This study aimed to isolate bacteria for the production of valuable bioactive compounds with antimicrobial activity from the compost of green waste mixed with phosphate residues and straws. Pseudomonas aeruginosa CFSP1L1 was identified as the most efficient bacterium in nine isolated strains in terms of inhibitory effect against Salmonella spp., Streptococcus spp., and Escherichia coli (E. coli). This bacterium has also been shown to have antifungal activity against Fusarium oxysporum f. sp. albedinis (Foa) and Aspergilus niger (A. niger). Its antimicrobial activity was not abolished after the heat and enzymatic treatments, showing that P. aeruginosa CFSP1L1 acts by bioactive substances of a non-protein, non-lipid and non-saccharidic nature. In vivo, the treatment of no damaged apples and mandarins with a suspension of the antagonist P. aeruginosa CFSP1L1 (108 CFU/ml) for 24 h and then inoculated with a suspension (107 CFU/ml) of A. niger and Foa, respectively showed that the bacterial strain CFSP1L1 completely inhibited the phytopathogenic effect of both fungi.
Keywords: Compost; Phytopathogens; Pseudomonas aeruginosa
Introduction
Agriculture plays a crucial socio-economic role in different countries. In Morocco, the agricultural sector is one of the main pillars of the national economy. For example, it contributes by 14 to 15% of the national gross domestic product [1]. However, it is exposed to several anthropogenic and/or natural constraints and in particular, those caused by pathogens, thus, leading to significant economic losses [1,2]. Fusarium is one of these agents, and it causes Fusarium wilt, a disease that affects cereals (wheat, barley, and other small grains) [3]. Species belonging to this genus also attack others vegetables and plants. For example, the case of Fusarium oxysporum f.sp. cucumerinum, which devastates cucumber [4] and Foa, which causes Fusarium wilt commonly known as bayoud. This is the most destructive fungal disease of the date palm [5]. Another example to cite is that of A. niger responsible for developing black mold of several organic substrates including fruits, vegetables, grains, seeds, spices, grass and wood. In addition, this fungal species can produce my cotoxins, which exhibit several adverse effects on the liver, kidneys, nervous system, muscles, digestive tract, respiratory and genital organs [6]. In addition to fungi, some bacteria can also infect plants. Among them, Salmonella which infects Arabidopsis by multiplying in plant tissue and thus leading to various symptoms such as wilting and yellowing of leaves [7]. On the other hand, many human disease outbreaks caused by food-borne pathogens have been associated with the consumption of contaminated fresh produce, such as alfalfa sprouts, lettuce, spinach, parsley, and cantaloupes [7].
Currently, the control of these phytopathogens is based on the use of pesticides. However, this chemical treatment has a toxic effect on human health and sometimes causes irreversible environmental damage, such as the contamination of surface and groundwater, soil impoverishment and reduction of biodiversity. Faced with this situation, farmers must adopt other alternatives without environmental or health threatening. In this sense, many bacteria resulting from composts of different organic wastes are used in agriculture as biological control agents. They can so replace harmful chemicals by improving the health of plants and by limiting the saprophytic growth of phytopathogenic microorganisms. Indeed, the investigations on biocontrol using microorganisms are experiencing remarkable growth. They are based on the mechanisms of microbial antagonism involved in the protection of plants against their enemies.
Among these microorganisms, P. aeruginosa, a Gram-negative bacterium can colonize many ecological niches including soil, composts, plants and aquatic surfaces [8]. The proliferation of the P. aeruginosa population provides information on the proper conduct of composting and the metabolization of organic waste material, especially in the final composting phase [9,10]. In fact, P. aeruginosa is considered to be thermo-tolerant during composting [11] and its proliferation in the final phase of composting has been reported by several authors [11,12].
Indeed, P. aeruginosa secretes various metabolites including pyocyanin, a blue-green pigment. Its antimicrobial effect against pathogenic microorganisms has been demonstrated [13]. Generally, many bacteria belonging Pseudomonas genus produce many antifungal metabolites such as phenazines, pyoluteorin, pyrrolnitrine, and DAPG (2,4- diacetylphloroglucinol), which are the most frequently detected antifungals [14-16]. These bacteria are also able to synthesize siderophores called pyoverdins or pseudobactins. These molecules are involved in improving the growth and health of plants [11] and contribute to the acquisition of iron by plants [10].
In this context, this present work consisted of the isolation of bacteria secreting compounds that had antibacterial and antifungal effect from a compost of green waste mixed with phosphate residues. Therefore, the antimicrobial activity of isolates was evaluated, as well as in vivo testing of the antagonistic effect of the best performing isolate against Foa and A. niger, and the partial characterization of secreted bioactive substances.
Materials and Methods
Sampling
To isolate microorganisms with an antimicrobial effect from the compost of green waste mixed with phosphate residues and straws elaborated as described. Three representative samples have been taken from the composts during the succession of three distinct phases: (1) mesophilic phase, (2) thermophilic phase, and (3) maturation phase.
Antimicrobial effect against pathogenic microorganisms
Pathogenic strains used and culture media: E. coli CCMM/B4, Streptococcus sp. CCMM/ B24 and Salmonella sp. CCMMB17 strains were used as tests to demonstrate antibacterial effects. The Foa A27 and A. niger CCMM- M100 were used as tests for antifungal effect. These fungal are grown in Potato Extract Glucose Agar (PDA). The dishes were incubated in an oven at 28 ± 1ºC for seven days, and then they were stored at 4ºC.
Meanwhile, the other culture media used are suitable for developing the target bacterial strains, and the majority of microorganisms are isolated from compost (fast and slow-growing microorganisms). Luria-Bertani (LB) medium issued for suspension cultures of pathogenic bacteria, and for inoculation of isolates of interest to produce bioactive substances.These bacteria were maintained in liquid LB medium (10 g of peptone, 5 g of yeast extract, 10 g of NaCl per one liter of distilled water), and solid LB medium (10 of peptone, 5 g of yeast extract, 10 g of NaCl and 15 g of agar per one liter of distilled water). During the entire period of this work, the bacteria were inoculated every week on solid media and then they were stored at 4ºC.
Isolation and screening of microorganisms producing substances with an antibacterial effect: The compost sample (10 g) was diluted in 90 ml of a buffer solution (0.06 M Na2HPO4/NaH2PO4) (1/9 v/v), pH 7.6. Dilutions of decimal series (10-1 to 10-7) of each sample of the composts were carried out. An aliquot of 100 μl of each dilution was then spread on a agar plates previously inoculated with E. coli, Salmonella sp. and Streptococci sp. After incubation at 37ºC for 48 hours, colonies surrounded by an inhibition halo were subculture and purified [2,17].
The antibacterial activity of the isolates: This test consisted of confirming the antibacterial effect of the isolates directly on a solid medium. Thus, a colony of each isolate was taken from a fresh culture, then placed in the centre of a plate containing solid LB medium previously inoculated with E. coli, Salmonella sp., and Streptococci sp. The dishes were incubated at 37ºC for 48 h. The diameters of the inhibition halos were measured according to [2]. The results were expressed by measuring the distance between the border of the bacterial colony and the start of the zone of inhibition of the pathogenic strain.
Antifungal effect of the isolate CFSP1L1 against A. niger and FoA: The antagonism of the isolate CFSP1L1 against the two phytopathogens (Foa and A. niger) was studied side-by-side co- culture on the solid PDA medium according to [5]. Thus, a Petri dish was streaked with the isolate. After 48 h of incubation, two 5 mm discs of a two days old culture of Foa were deposited 2.5 cm from the streak of the isolate against each other perpendicular to the line of the seeding streak. The petri dish was then incubated at 25 ± 2ºC. The percentage inhibition was evaluated by measuring the radius of the colony of Foa co-cultivated with the isolate CFSP1L1. The radius of the colony of a control cultivated in the absence of any other microorganism was also evaluated. Measurements were taken after 6 days of incubation. The same procedure was followed to test the antagonistic effect of the isolate against Aspergilus niger. For the validation of the experimental protocol carried out, 3 repetitions were made for both pathogenic agents (FoA and A. niger).
Identification of the isolate CFSP1L1 that produces substances with antimicrobial effect
Total DNA extraction: The total DNA of the bacterial isolate was obtained by phenol-chloroform extraction from exponentially growing bacterial cultures [18]. The cells were centrifuged by centrifugation at 13,000 rpm for 5 min, then suspended in lysis buffer (40 mM Tris- acetate, pH 7.8, 20 mM sodium acetate, 1 mM EDTA, 1% SDS, 20 µg mL-1 RNAse). Then 100 µl of 5 M NaCl was added to the solution, the tubes were centrifuged at 13,000 rpm for 10 min at 4ºC. The supernatants were transferred to new microtubes and equal volumes of phenol- chloroform: isoamyl alcohol (25: 24: 1, v/v) were added. The DNA was precipitated in absolute ethanol and washed twice with 70% ethanol, then dried and redissolved in pure sterile water. The DNA's concentration and purity were evaluated using a Nano Drop TM spectrophotometer and was stored at -20ºC for later use.
Amplification of DNA encoding 16S rRNA: The DNA encoding the 16S rRNA of the isolate of interest was amplified using the two primers rD1(5'AAGGAGGTGATCCAGCCGCA3') and fD1 (5'GGAGAGTTAGATCTTGGCTC3') (Weisburg et al.), in a reaction volume of 25 μl, the mixture containing: 1 μl of DNA with a concentration of 100 ng/μl, 1 μl of each primer at 10 μM, 9.5 μl of pure sterile water and 12.5 μl of MytTaqTM Mix, 2x ( BIOLINE) ready to use, containing the Taq polymerase enzyme, reaction buffer, MgCl2 and dNTPs. The following cycles were used under the optimal conditions supplied with the MytTaqTM Mix: initial denaturation (95ºC for 60 seconds), followed by 35 cycles (95ºC for 15 seconds, hybridization temperature 60ºC for 15 seconds, 72ºC for 10 seconds), and followed by 1 min final extension at 72ºC.
Sequencing and phylogenetic analysis of the 16S rRNA gene: The 16S rRNA amplification products were purified using the Quiagen PCR product purification system, then subjected to cycle sequencing using the same primers as for PCR amplification, with ABI Prism Dye Chemistry, and analysed with a 3130xl automatic sequencer [19], in the sequencing facilities of the Innovation city in Fez (Morocco). The nucleotide sequences obtained were compared with those of the reference strains available in the Gen-Bank database (NCBI: National Center for Biotechnology Information), using the Blast software (Basic Local Alignment SearchTool (http: / www. ncbi.nlm.nih.gov /Blast.cgi) (20) The nucleotide sequences were aligned, verified and corrected manually using Chromas 2 (Version 2.6.5) and MEGA 7 (Version 7.0.26). The sequences obtained were deposited and identified in the Genbank database. The obtained accession number of the isolated strain was represented in Table 2.
In vivo effect on A. niger and Foa
Apples and mandarins were washed thoroughly with sterile distilled water before use for inoculation with Isolated strain CFSP1L1 using the protocol of [20,21]. Briefly, the antagonist's bacterial suspension was prepared in sterile distilled water and was adjusted to 108 CFU/ml. Injured apples and mandarins has been injected with a suspension of the Isolated strain CFSP1L1 (108 CFU/ml) for 24 h and then inoculated with a suspension of A. niger (107 CFU/ml) and Foa, respectively. The results were recovered after 15 days post inoculation. Apples and mandarins injected only with the antagonist were used to assess the cytotoxicity of the interest isolate. While apples and mandarines treated with distilled water or with the phytopathogen fungi A. niger or Foa were used as controls
Preliminary characterization of substances with antibacterial activity produced by the isolated strain CFSP1L1
Extraction of bioactive substances by ethyl acetate: The precipitation of bioactive substances from the isolate was carried out according to Naclerio's protocol and his collaborators (1993). Thus, a volume of 100 ml of sterile liquid LB was inoculated with the bacterial suspension, and then incubated with shaking at 37ºC for two days.
After centrifugation of this culture at 5000 rpm for 15 min at 4ºC, the supernatant was recovered and then added to 100 ml of organic solvent (ethyl acetate at 95% purity). The mixture was stirred for two hours to facilitate the extraction of the secreted bioactive compounds.
The product was decanted in the separating funnel for one hour followed by the elimination of the aqueous phase and recovery of the organic phase. After then, the evaporation of ethyl acetate was carried out via a rotary evaporator. The precipitated compounds were solubilized in 1 ml of sterile distilled water [22].
Testing of the antibacterial activity of the bioactive substances by the well method: The well technique consists of distributing the LB Agar medium in Petri dishes. After solidification, wells of identical volume were made using a sterile pasteur pipette. Then, a volume of 100 μl of the extract of the Isolated strain CFSP1L1 was incubated at 37°C for 3 hours, and then tested according to the well method on dishes previously inoculated with E. coli; Salmonella sp and Streptococci sp. The dishes were incubated at 37ºC for 48 hours then the diameters of the inhibition zones were measured [2,23].
Nature of the bioactive substances secreted by the isolate of interest:
Sensitivity to proteinase K: In order to determine whether the bioactive compounds are of a protein nature, the extracts precipitated by ethyl acetate were subjected to the action of proteinase K (PK) at a concentration of 1 mg/ml at a pH 7. The mixture was well homogenized and incubated at 37ºC for 3 hours.
The antibacterial effect of the bioactive compounds treated with PK was studied against E. coli using the well method. Petri dishes were incubated at 37ºC for 48 hours [1]. Extracts not treated with PK were used as controls.
Sensitivity to lipase: To demonstrate whether the bioactive compounds are lipidic in nature, the precipitates were subjected to lipase action at a concentration of 1 mg/ml at a pH 7. The mixture was well homogenized and incubated at 37ºC. for 3 hours. The antibacterial effect of the bioactive compounds treated with lipase was studied against E. coli using the good method. Petri dishes were incubated at 37ºC for 48 hours. Extracts not treated with lipase were used as controls [2].
Sensitivity to amylase: In order to determine whether the bioactive substances are of carbohydrate nature, the various precipitates were subjected to the action of amylase at a concentration of 1 mg/ml at a pH 7. The mixture was well homogenized and incubated at 28ºC during 3 hours.
The antibacterial effect of the bioactive substances treated with amylase was studied against E. coli using the well method. Petri dishes were incubated at 37ºC for 48 hours. Extracts not treated with amylase were used as controls [2].
Sensitivity to heat treatment: To test the sensitivity of the bioactive substances to heat treatment, the protocol of Naclerio, 1993 is adopted. The precipitated extracts with ethyl acetate were exposed to 100ºC (15 min), 80ºC (15 min), 60ºC (20 min) and 37ºC (3 h). The treated precipitates were tested against E. coli using the well method. The dishes obtained were incubated at 37ºC for 48 hours.
Results and Discussion
Isolation and screening of microorganisms producing bioactive substances
Composting is naturel process based on the development of intense microbial activities leading to the decomposition of most bioavailable and biodegradable materials. This biological process involves the partial or complete degradation of various chemical compounds by a variety of microorganisms [24,25].
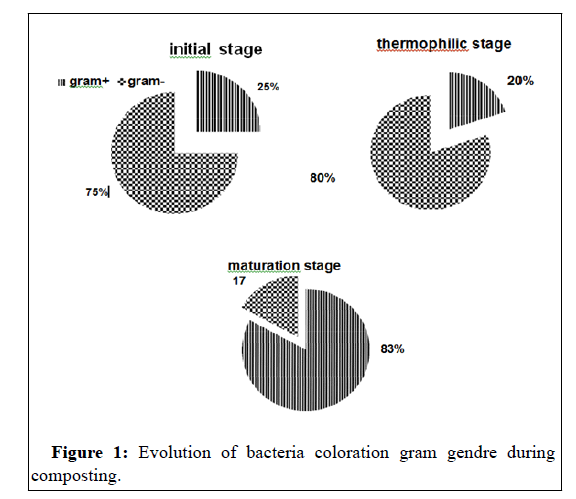
During the composting phases, the microbial community follows a predictable succession pattern that results in the recolonization of the compost with metabolically active microbial populations that can be suppressive to phytopathogens. In this compost, as demonstring in previous studies [9,26],the gram+bacteria become the most predominant in maturation step, while the gram- negative that dominant in the initial wastes has been eliminated (Figure 1).
In fact, the production of antimicrobial agents, competition for nutrients, oxygen or space is the mechanisms that allow the microbial population in a compost to control or inhibit the effect of disease-causing pathogens [18].
Indeed, the suppression of several plant diseases by the activity of microorganisms from compost has been described [27]. It is in this sense that samples of the compost of green waste mixed with phosphate residues during the mesophilic, thermophilic, and maturation phases has been used to isolate a promoting microorganism producing substances that inhibit E. coli; Salmonnella sp and Streptococci sp. Among bacteria, ten were isolated with inhibitory potential. Indeed, these isolates showed halos of inhibition of different diameters around the colonies of the above three pathogens (Table 1). Some of these microorganisms have a broad spectrum. While others have a narrow spectrum acting only on a single target bacteria. All of these isolates act with one or more bioactive substances that inhibit bacterial proliferation. In this work, the choice has been made on the CFSP1L1 isolated from the compost's maturation phase. Indeed, it exhibits an interesting antibacterial effect by showing halos of inhibition of order 15, 16 and 18 mm towards Streptococcus sp, Salmonella sp and E. coli, respectively. This agrees with some previous research, which suggests the pathogen suppressor effect of composts isolates [27,28]. [27] Have demonstrated the antimicrobial activity of Pseudomonas, Serratia, Klebsiella, Enterobacter against specific pathogens of turf, namely Sclerotinia homoeocarpa, Pythium graminicola, Typhulai shikariensis, and Microdochium nivale. In another study performed in vitro by [29], it was found that compost contains 44 species of fungi and 15 species of bacteria, of which seven belong to three genera namely Bacillus, Micrococcus and Pseudomonas. These bacteria have been shown to have an inhibitory power against pathogens [29]. These authors have demonstrated that these microorganisms produce bioactive molecules that suppress pathogens through their metabolisms' secondary pathways or spontaneous expression of genes induced by pathogens.
| Isolates | Compost phase from which the isolate is obtained | Inhibition diameters in (mm) against | ||
|---|---|---|---|---|
| Streptococcus sp | Salmonella sp | E. coli | ||
| 2CFE51 | Maturation phase | - | 6 | - |
| 2CTE51 | Thermophilic phase | 5 | 8 | - |
| CAFE32 | Maturation phase | 8 | 22 | 19 |
| CFSL32 | Maturation phase | 20 | 7 | 13 |
| CFSP1L1 | Maturation phase | 15 | 16 | 18 |
| CFSPL12 | Maturation phase | 6 | - | 7 |
| CIca71 | Mesophilic phase | 7 | 5 | - |
| CISP31 | Mesophilic phase | 6 | 7 | 9 |
| CTSL31 | Thermophilic phase | 40 | - | - |
| CTSPL32 | Thermophilic phase | 10 | 19 | 11 |
- : absence of inhibition
Table 1: Antibacterial activity of isolates from compost of green waste mixed with phosphate residues.
Antifungal effect of the isolate CFSP1L1 against A. niger and Fao
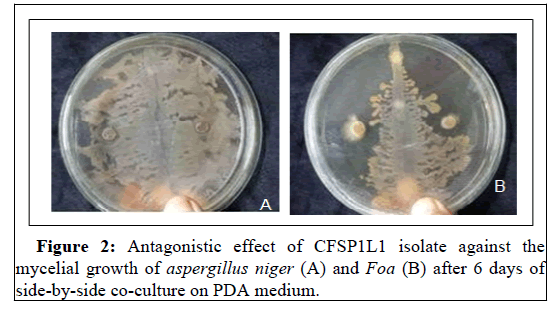
In vitro tests show that the CFSP1L1 isolate exhibits a significant effect of inhibiting the sporulation of A. niger and Foa in the order of 99% and 90%, respectively (Figure 2). The suppressant effect of the compost on pathogens including Fusarium oxysporum f. sp has already been shown by [30]. In fact, compost probably acts as much through its intrinsic microbial activity by stimulating the soil's microbial activities [31]. In addition, the addition of compost increases the suppressive effect against Fusarium oxysporum f .sp. This effect is proportional to the amount of compost administered [32].
Identification of the isolate CFSP1L1
The CFSP1L1 isolate is characterized by colonies of brown color, rounded relief, smooth and shiny appearance as well as a particular aromatic odor. Gram stain revealed that it is a mobile Gram-negative bacillus, arranged singly or in pairs. This result is consistent with that of the molecular identification of the isolate. Indeed, after purification of the PCR products, the DNA encoding 16S rRNA was sequenced using the primers rD1 and fD1. Sequencing of 16S rDNA resulted in a sequence having 1439 nucleotides. In the literature, identifications have been made using sequences of 500 bp [33,34] and others have been based on the sequencing of about 400 bp [35].
Accordingly, after bioinformatics analysis, the bacterial specie whose 16S rRNA gene shows a strong identity of around 99.79% with that of the isolate is P. aeruginosa (Table 2). Basing on the criteria defined by [36], this percentage allows identifying the isolate. An accession number was provided to the sequence obtained after being deposited in the Genbank database (MT415324). Thus, based on Gram stain and molecular identification, the isolate CFSP1L1 isolate has been assigned to P. aeruginosa.P. aeruginosa is a ubiquitous bacterium that colonizes soil, plants, aquatic surfaces as well as composts [8,37]. Indeed, it is considered to be thermo-tolerant during composting [11] and its proliferation in the final phase of composting has been reported by several authors [10,12,27]. This makes it possible to explain its isolation from the compost maturation phase during this present work. Nevertheless, to date, the antimicrobial effect of P. aeruginosa isolated from compost has not yet been strictly demonstrated. [38] Has suggested that P. aeruginosa is one of the microorganisms in compost responsible for a significant reduction in the microbial population, including the genus Salmonella. Indeed, various studies have shown that the bacteria in question have a broad spectrum of action against Gram-positive and Gram-negative bacteria as well as against fungi. The investigation by [1] has reported that the said bacteria inhibit the growth of E. coli. Likewise, in the study performed by [39], it was reported that P. aeruginosa may limit the growth of Streptococcus spp through several mechanisms including surfactant and alginate synthesis as well as production of siderophores via iron sequestration.
| Isolate (Accession number) | 16S rRNA sequence size identity percentage | Type strain showing strong similarity to the isolate |
|---|---|---|
| CFSP1L1 (MT415324) 1439n | 99,79% | Pseudomonas aeruginosa ATCC 10145 (NR_114471) |
Table 2: Results of molecular identification of isolate CFSP1L1 after sequencing of the 16S rRNA gene.
On the other hand, it has been shown that P. aeruginosa is effective in controlling peanut crown rot disease caused by A. niger by inducing hyphal deformities in the fungal pathogen [40]. In addition, protection against Fusarium oxysporum f. sp by fluorescent Pseudomonas has also been revealed by several authors [4,41-43] corroborates the findings of the present investigation. Indeed, the current work is the first to demonstrate that P. aeruginosa has an antifungal effect against Foa. This therefore makes P. aeruginosa an excellent candidate to exploit in order to control phytopathogens.
Antagonistic effect in vivo of P. aeruginosa against A. niger and Foa
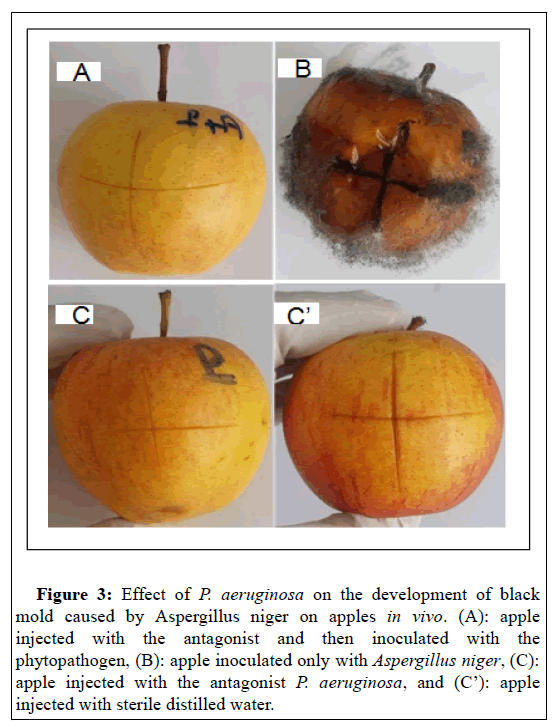
The treatment of apples with a suspension of the antagonist P. aeruginosa (108 UFC/ml) for 24 h and then inoculated with a suspension of the pathogen (107 UFC/ml) shows that P. aeruginosa completely inhibits the appearance of black mold due to A. niger after 15 days post inoculation (Figure 3A). In contrast, the apples were completely rotten after their individual inoculation with a suspension of the fungal species at 107 UFC/ml (Figure 3B). As control, inoculation of apples with the antagonist P. aeruginosa at a rate of 108 UFC/ml does not reveal symptoms of pathogenicity to the plant tissue tested (Figure 3C). The same result was seen for the distilled water control (Figure 3C).
Figure 3: Effect of P. aeruginosa on the development of black mold caused by Aspergillus niger on apples in vivo. (A): apple injected with the antagonist and then inoculated with the phytopathogen, (B): apple inoculated only with Aspergillus niger, (C): apple injected with the antagonist P. aeruginosa, and (C’): apple injected with sterile distilled water.
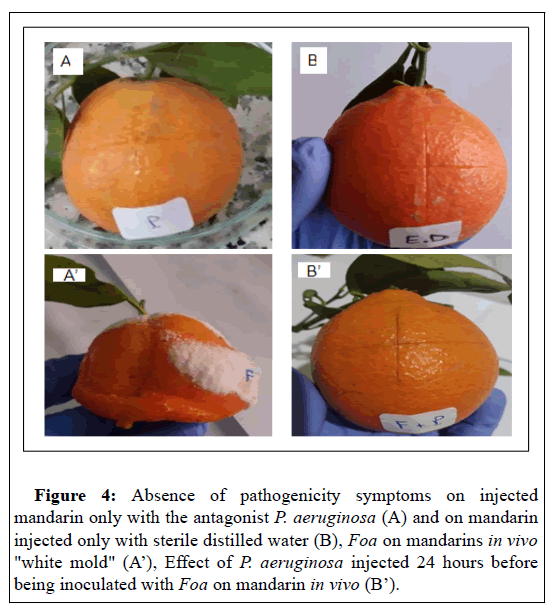
Therefore, P. aeruginosa could be used to confer protection on injured apples after harvest against A. niger. This finding can be explained by the fact that the bacteria secrete metabolites that inhibit fungal growth. Indeed,[44] have reported that the cell- free culture filtrate of P. aeruginosa GSE 18, inhibited in vitro the production of polygalacturonase and cellulase by A. niger six days after inoculation, by up to 77% and 68%, respectively. Such enzymes break down pectin and cellulose, the two main polymers that maintain the firmness and structure of plant cell walls. Subsequently, the degradation of the structural molecules of the wall allows the phytopathogen to penetrate and colonize the plant tissue. In fact, inhibition of these enzymes' production represents a mechanism that may have an essential role in controlling decay by reducing the virulence of A. niger [44]. Also, inoculation of mandarins of Citrus reticulata Blanco species with the antagonist P. aeruginosa, 24 before Foa completely inhibits the occurrence of white sporulation due to Foa compared to controls (Figure 4). The antagonist P. aeruginosa isolated in this present work could act by a similar mechanism to inhibit the plant pathogen's growth. However, to validate this hypothesis, investigations must be carried out to reveal and fully understand the mechanism of action of the antifungal effect of P. aeruginosa.
Figure 4: Absence of pathogenicity symptoms on injected mandarin only with the antagonist P. aeruginosa (A) and on mandarin injected only with sterile distilled water (B), Foa on mandarins in vivo "white mold" (A’), Effect of P. aeruginosa injected 24 hours before being inoculated with Foa on mandarin in vivo (B’).
Antibacterial activity of the bioactive substances of P. aeruginosa
The extraction of the bioactive substances from the CFSP1L1 isolate was carried out using ethyl acetate as an organic solvent. Such a solvent is widely used to extract metabolites from bacteria [1,45]. After extraction, the antibacterial effect of the metabolites of P. aeruginosa was evaluated by the well method. Thus, the results show that the extract has an antibacterial activity that has been demonstrated by the appearance of inhibition halos. The diameters of the later are of the order of 17 mm, 18 mm and 20 mm towards Streptococcus sp, Salmonella sp, and E. coli, respectively. This result indicates that P. aeruginosa acts by antibacterial metabolites secreted in the LB culture medium and also highlights that these substances are soluble in ethyl acetate.
Stability and nature of the bioactive substances of P. aeruginosa
In order to better elucidate the stability and chemical nature of the bioactive molecules secreted by P. aeruginosa, the extract has been subjected to several treatments. Its antibacterial activity was then evaluated against E. coli by the excellent technique. The results show that the antagonistic effect of CFSP1L1 was not affected by heat or by enzymatic treatments with PK, lipase and amylase. This implies that the bioactive compound responsible for the desired activity are not protein, lipid or saccharide (Table 3)
| Enzymatic treatment | Relative activity |
|---|---|
| Protéinase K | + |
| Lipase | + |
| Amylase | + |
| Control (untreated ethyl acetate extract) | + |
| Heat treatment | |
| 37ºC | + |
| 60ºC | + |
| 80ºC | + |
| 100ºC | + |
| Control (untreated ethyl acetate extract) | + |
Relative activity was measured by the well method against E. coli.+ : presence of inhibition
Table 3: Effect of the different treatments on the antibacterial effect of P. aeruginosa extract.
Previously, P. aeruginosa has been reported to produce a variety of metabolites including phenazines. These molecules represent a large family of highly pigmented nitrogenous heterocyclic molecules synthesized from shikimic acid [46,47]. As an example of these molecules exhibiting antimicrobial activities, it has been reported the molecules such as carboxylic acid 1 phenazine, phenazine 1 carboxamide and pyocyanin [2,37]. The latter metabolite is a very distinctive blue pigment produced only by P. aeruginosa and is soluble in organic solvents [2]. Purified pyocyanin has been shown to have significant antimicrobial activity against pathogens tested such as Staphylococcus aureus, E. coli, Candida albicans, Bacillus cereus and Salmonella sp [13]. In addition, several studies have shown that this bioactive substances is stable at high temperature and it shows resistance to boiling without losing its antibacterial effect [1,2,46-48].
In addition, phenazines are biologically active having a role in microbial competitiveness and suppressing plant pathogens [2]. In fact, phenazines have been shown to inhibit the growth of phytopathogenic fungi such as Gaeumannomyces graminis, Fusarium oxysporum, Rhizoctonia solani and Gibbbereala avenacea [49,50]. Consequently, the bioactive substances secreted by P. aeruginosa could be organic compounds in the form of phenazines [51,52]. Thus, more work needs to be done to further purify and elucidate the chemical structure of the bioactive substances in the isolate [53,].
Conclusion
Post-harvest plant and agricultural product diseases caused by microorganisms cause serious economic losses internationally. Thus, biological control is experiencing remarkable growth as an alternative to conventional treatment based essentially on the use of pesticides. Mainly of that, the excessive use of the later has generated the emergence of pathogens resistant to chemical molecules and them also shown harmful effects on the environment, human health. It is in this sense that P. aeruginosa was isolated, during this present work, from a compost of green waste mixed with phosphate residues. This microorganism has an interesting antibacterial effect against Gram positive and Gram-negative bacteria and also an antifungal activity against Foa and A. niger. In addition, P. aeruginosa has been shown to completely inhibit in vivo the appearance of black mold due to A. niger and Foa after 15 days post inoculation of apples. More interestingly, it does not manifest symptoms of pathogenicity with respect to the plant tissue tested. Therefore, it appears that P.aeruginosa is a promising strain which deserves to be exploited as a potential antagonistic agent in order to confer post-harvest protection to plants against biotic threats. To achieve this objective, other additional investigations seem necessary to be carried out such as the field application of the strain as well as the purification of its bioactive substances.
References
- Zahir I, Babouchi M, Boulanour H, Mustapha ELL (2018) Effet des microorganismes isolés á partir des biotopes marocains sur les phytopathogènes : revue bibliographique Revue Agrobiologia . 8(2): 971-983.
- Zahir I, Houari A, Iraqui M, Ibnsouda S (2018)Valorisation de l’activité antibactérienne des microorganismes isolés à partir des biotopes marocains et caractérisation partielle de leurs principes actifs. 80-95.
- Nitschke M, Aeauujo LV, Costa SGVAO, Pires RC, Zeraik AE, et al. (2009) Surcaftin reduces the adhesion of food-borne pathogenic bacteria to solid sufaces. Lett Appl Microbiol 49: 241-247.
- Islam MDA, Nain Z, Alam MDK, Banu NA, Islam MDR (2018) In vitro study of biocontrol potential of rhizospheric Pseudomonas aeruginosa against Fusarium oxysporum f. sp. Cucumerinum. Egypt J Biol Pest Control 28(90).
- Hassni MEI, Hadrami AEI, Daayf F, Chérif M, Barka EA, et al.(2007) Biological control of bayoud disease in date palm: Selection of microorganisms inhibiting the causal agent and inducing defense reactions. Environ Exp Bot 59(2): 224-234.
- Â Gautam AK, Sharma S, Avasthi S, Bhadauria R (2011) Diversity, pathogenicity and toxicology of A. niger: An important Spoilage Fungi. Res J Microbiol 6(3): 270-280.
- Gautam AK, Sharma S, Avasthi S, Bhadauria R (2011) Diversity, pathogenicity and toxicology of A. niger: An important Spoilage Fungi. Res J Microbiol 6(3): 270-280.
- Magin V (2019) Exploitation du potentiel des bactériophages dans le traitement des surfaces en contact avec l’eau, contaminées par un biofilm de Pseudomonas aeruginosa.
- Seo S, Matthews KR (2012) Influence of the Plant Defense Response to Escherichia coli O157:H7 Cell Surface Structures on Survival of That Enteric Pathogen on Plant Surfaces. Appl Environ Microbiol 78(16): 5882-5889.
- Amir S, Merlina G, Pinelli E, Winterton P, Revel JC, et al. (2008) Microbial community dynamics during composting of sewage sludge and straw studied through phospholipid and neutral lipid analysis. J Hazard Mat 159(2-3): 593-601.
- Lemanceau P, Expert D, Gaymard F, Bakker PAHM, Briat JF (2009) Role of iron in plant–microbe interactions. Adv Bot Res 51: 491-549.
- Atif K, Haouas A, Aziz F, Yasser Jamali MY, Tallou A,et al. (2020) Pathogens evolution during the composting of the household waste mixture enriched with phosphate residues and olive oil mill wastewater. Waste and Biomass Valorization 11(5): 1789-1797.
- Barakat R (2012) Etude des propriétés biologiques et antimicrobiennes de la pyocyanine, pigment redox-actif produit par Pseudomonas aeruginosa. Sciences agricoles.
- Dieng M, Diedhiou AS, Sambe FM (2019) Valorisation par compostage des déchets solides fermentescibles collectés à l’Ecole Supérieure Polytechnique de l’Université Cheikh Anta Diop de Dakar: Etude de l’effet phytotoxique sur des plants de maïs et d’arachide. IJBCS 13(3): 1693-1704.
- Haas D, Défago G (2005) Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbial 3(4): 307-319.
- Weller DM, Landa BB, Mavrodi OV, Schroeder KL, de la Fuente L, et al. (2007) Role of 2,4- diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in the defense of plant roots. Plant Biol 9: 4-20.
- Weller DM, Raaijmakers JM, McSpadden-Gardener BB, Thomashow LS (2002). Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu Rev Phytopathol 40: 309-348.
- Hassi M, Haggoud A, Mzibri MEI, Ibnsouda S, Houari A, et al. (2007) Isolation and identification of a staphylococcal strain with an anti-mycaobacterial activity and study of it’s mode of action. Ann Microbial 57(4): 651-656.
- Haouas A, Modafar CEI, Douira A, Ibnsouda-Koraichi S, Filali-Maltouf A, et al. (2021) Alcaligenes aquatilis GTE53: Phosphate solubilising and bioremediation bacterium isolated from new biotope “phosphate sludge enriched- compost. Saudi J Biol Sci 28(1): 371-379.
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74(12): 5463-5467.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3): 403-410.
- Bahadou SA, Ouijja A, Karfach A, Tahiri A, Lahlali R (2018). New potential bacterial antagonists for the biocontrol of fire blight disease (Erwinia amylovora) in Morocco. Microb Pathog 117: 7-15.
- Guendouzi SEI (2014) Isolement et identification de bactéries sécrétant des substaces à effet antimycobactérien, caractérisation partielle des métabolites bioactifs et étude préliminaire de leur mode d’action. Thèse de 3eme cycle, Université Sidi mohamed ben abdellah Faculté des sciences et techniques Fès Maroc.
- Amir S (2005) Contribution à la valorisation de boues de stations d’épuration par compostage devenir des micropolluants métalliques et organiques et bilan humique du compost.
- Zahir I, Houari A, Ibnsouda S (2014) Antibacterial Effect of Pseudomonas aeruginosa Isolated from a Moroccan Hot Spring Discharge and Partial Purification of its extract. J British Biotechnology 4(10): 1123-1140.
- Amir S, Abouelwafa R, Meddich A, Souabi S, Winterton P, et al. (2010) PLFAs of the microbial communities in composting mixtures of agro-industry sludge with different proportions of household waste. Int Biodeterior Biodegradation 64(7): 614-621.
- Haouas A, Modafar CEI, Douira A, Ibnsouda-Koraichi S, Filali-Maltouf A, et al. (2021) Evaluation of the nutrients cycle, humification process, and agronomic efficiency of organic wastes composting enriched with phosphate sludge. J Clean Prod. 302: 127051.
- Boulter JI, Trevors JT, Boland GJ (2002) Microbial studies of compost: bacterial identification, and their potential for turfgrass pathogen suppression. World J Microbiol Biotechnol 18: 661-671.
- Ramzan NR, Shahzad S (2014) Inhibition of in vitroGrowth of soil-borne pathogens by compost inhabiting indigenous bacteria and fungi. Pak J Bot 46(3): 1093-1099.
- Larbi M (2006) Influence de la qualité des composts et de leurs extraits sur la protection des plantes contre les maladies fongiques, Thèse présentée à la Faculté des Sciences de l’Université de Neuchâtel. Suisse.
- Nelson EB, Michel JB (2002) Microbial Mechanics of Compost-Induced Disease Suppression. Compost users forum.
- Craft CM, EB Nelson (1996) Microbial properties of composts that suppress damping- off and root rot of creeping bentgrass caused by Pythium graminicola. Appl Environ Microbial 62(5): 1550-1557.
- Serra-Wittling C, Sabine H, Enrique B (1996) Modification of soil water retention and biological properties by municipal solid waste compost. Compost Sci Util 4(1): 44-52.
- Hall L, Doerr KA, Wohlfiel LS, Roberts GD (2003) Evaluation of the MicroSeq system for identification of mycobacteria by 16S ribosomal DNA sequencing and its integration into a routine clinical mycobacteriology laboratory. J Clin Microbiol 41(4): 1447-1453.
- Patel JB, Leonard DG, Pan X, Musser JM, Berman RF, et al. (2000) Sequence- based identification of Mycobacterium species using the Microseq 500 16S rDNA bacterial identification system. J Clin Microbiol 38(1): 246-251.
- Bosshard PP, Abels S, Zbinden R, Bottger EC, Altwegg M (2003) Ribosomal DNA sequencing for identification of aerobic Gram-positive rods in the clinical laboratory (an 18 month evaluation). J Clin Microbiol 41(9): 4134-4140.
- Rane MR, Sarode PD, Chaudhari BL, Chincholkar SB (2007) Detection, isolation and identification of phenazine-1-carboxylic acid producer by biocontrol strains of Pseudomonas aeruginosa. J Sci Ind Res 66(8): 627-631.
- Rodnguez C (1996) Viviana Beoletto and M6nica Finola bacteriology of poultry litter, compost and the earthworm Eisenia foetida (oligochaeta, lumbricidae) megadrilogica. 6(10).
- Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral JP, et al. (2000) 16S Ribosomal DNA Sequence Analysis of a Large Collection of Environmental and Clinical Unidentifiable Bacterial Isolates. J Clin Microbiol 38(10): 3623-3630.
- Scott JE, Li K, Filkins LM, Zhu B, Kuchma SL, et al. (2019) Pseudomonas aeruginosa can inhibit growth of streptococcal species via siderophore production. J Bacteriol 201(8): e00014-19.
- Kishore GK, Pande S, Podile AR (2005) Biological control of collar rot disease with broad spectrum antifungal bacteria associated with groundnut. Can J Microbiol 51(2): 123-132.
- Weller DM, Cook RJ (1986) Increased growth of wheat by seed treatments with fluorescent pseudomonads, and implications of Pythium control. Can J Plant Pathol 3: 328-334.
- Lemanceau P, Alabouvette C (1991) Biological control of Fusarium diseases by fluorescent Pseudomonas and non-pathogenic Fusarium. Crop Protec 10: 279-286.
- Saber FMA, Abdelhafez AA, Hassan EA, Ramadan EM (2015) Characterization of fluorescent pseudomonads isolates and their efficiency on the growth promotion of tomato plant. Ann Agric Sci 60(1): 131-140.
- Kishore GK, Pande S, Podile AR (2006) Pseudomonas aeruginosa GSE 18 inhibits the cell wall degrading enzymes of Aspergillus niger and activates defence-related enzymes of groundnut in control of collar rot disease. Australasian Plant Pathology 35: 259-263.
- Zahir I, Houari A, Iraqui M, Ibnsouda S (2011) Aerococcus sp. with an antimycobacterial effect. Afr J Biotechnol 10(83): 19473-19480.
- Dwivedi D, Johri BN (2003) Antifungals from fluorescent pseudomonads: Biosynthesis and regulation. Current Science 85(12): 1693-1703.
- Saha S, Thavasi R, Jayalakshmi S (2008) Phenazine pigments from Pseudomonas aeroginosa and their application as antibacterial agent and food colourants. Res J Microbiol 3(3): 122-128.
- Cohain N, Thomashow LS, Mavrodic DV, Blankenfeldt W (2006) The purification, crystallization and preliminary structural characterization of PhzM, a phenazine- modifying methyltransferase from Pseudomonas aeruginosa. Acta Crystallographica Section F : Structural Biology and Crystallization Communications 62: 887-890.
- Denning GM, Iyer SS, Reszka K, O’Malley Y, Rasmussen GT, et al. (2003) Phenazine-1-carboxylic acid, a secondary metabolite of Pseudomonas aeruginosa, alters expression of immunomodulatory proteins by human airway epithelial cells. Am J Physiol Lung Cell Mol Physiol 283(3): 584-592.
- Zahir I, Houari A, Iraqui M, Ibnsouda S (2014 ) Tolerance tests of Alcaligenes faecalis BW1 extract. British Microbiology Research Journal 4(8): 905-917.
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ(1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173(2): 697-703.
- Haouas A, Modafar CEI, Douira A, Ibnsouda-koraichi S, Filali-maltouf A, et al. (2020) The effect of phosphate and organic additives on the stability of food waste in the full-scale composting. PCBMB 21(39-40): 17-28.
- Barguigua A, Zahir I, Fikri N, Youss S, Youss B (2020) Prospection des maladies microbiennes de l’olivier dans la région Tadla-Azila. Revue Marocaine des Sciences Agronomiques et Vétérinaires 8(3): 1-13.
Citation: Amir S, Atif K, Haouas A, Zahir I, Tallou A, et al. (2021) Potential for Bio Control of Fungal Phytopathogens by Pseudomonas Aeruginosa CFSP1L1 Isolated from a Matured Compost Sample. JBRBD 12:004. DOI: 10.4172/2155-6199.1000004
Copyright: © 2021 Amir S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 3500
- [From(publication date): 0-2021 - Apr 03, 2025]
- Breakdown by view type
- HTML page views: 2774
- PDF downloads: 726