Research Article Open Access
Potential Application of Modified Saccharomyces Cerevisiae for RemovingLead and Cadmium
| Lívia de CF*, Mario HB and Benedito C | |
| Departamento of Microbiology, University of Sao Paulo, Sao Paulo, Brazil | |
| Corresponding Author : | Lívia de CF Lívia de Carvalho Fontes Departament of Microbiology-University of Sao Paulo, Brazil Tel: +55-11-30917295 E-mail: liviacfontes@gmail.com |
| Received September 08, 2015; Accepted October 07, 2015; Published October 10, 2015 | |
| Citation: Lívia de CF, Mario HB, Benedito C (2015) Potential Application of Modified Saccharomyces Cerevisiae for Removing Lead and Cadmium. J Bioremed Biodeg 6:313. doi:10.4172/2155-6199.1000313 | |
| Copyright: © 2015 Lívia de CF, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
| Related article at Pubmed, Scholar Google | |
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Recently, heavy metal pollution has become a worldwide concern. Saccharomyces cerevisiae have been used for bioremediation process for heavy metal uptake. In this study, S. cerevisiae was genetically modified with the gene EC20 (synthetic phytochelatin) with the purpose to enhance the accumulation capability for Lead (Pb2+) and Cadmium (Cd2+). S. cerevisiae has showed high biosorption capability of Pb2+, followed by Cd2+. Indeed, the transformed cells exhibited even higher capacity to accumulate lead. TEM (transmission electronic microscopy) images of transformed S. cerevisiae showed accumulation of heavy metals on cell wall, which was also verified by EDS (energy dispersive spectrum). There are many mechanisms involved in metal uptake and for some unknown reason. The results of yeast expressing EC20 were better detected when yeast cells were grown in the presence of Pb2+ than with Cd2+, as clearly observed with the improvement in Pb2+ retention by yeast cell wall.
| Keywords |
| Yeast; Genetic modification; Transformation; Heavy metals; Cadmium; Lead uptake |
| Introduction |
| In recent years, pollution of wastewater has increased and heavy metals (such as lead and cadmium) are one of the heavy metals in the environment, causing health effects [1-4] and serious environmental problems [5-8]. Mining, industrial and volcanic activities are the main source of heavy metals in environment [9]. In the aquatic system, heavy metal pollution has become an environmental concern, as it is extremely recalcitrant and non-degradable. Even at low concentrations, heavy metals can cause toxicity to humans and other forms of life [5]. It is necessary to uptake these heavy metals from wastewater before its disposal. The toxicity of heavy metals could be transformed from relevant low toxic species into more toxic forms. Moreover, most of the heavy metal salts are soluble in water, hindering some methods of separation (such as, chemical oxidation or reduction, electrochemical treatment, evaporative recovery, filtration, chemical precipitation, ion exchange, membrane technologies, adsorption on activated carbon) [10]. As a result, microbial biomass is an option for developing economic and eco-friendly wastewater treatment process [8,11]. An alternative process is called biosorption that can be defined as the removal of metal from solution by biological material (biosorbents) [12]. These biosorbents can effectively sequester dissolved metal ions of solutions with high efficiency and speed. It also offers several advantages over conventional treatment methods including cost effectiveness, minimization of chemical/biological sludge and regeneration of biosorbent with possibility of metal recovery [13]. Metal polluted environment contains fungi, which has adapted to toxic concentration of heavy metals and become metal resistant [14]. Metals retention by living fungi depends on several factors such as culture conditions and metal concentration [15]. Especially, fungi used for biosorption can be fulfilled by simple physical methods without any damaging the fungi structural integrity [16]. Saccharomyces cerevisiae can be easily cultivated using unsophisticated fermentation techniques and inexpensive growth media [17] at large scale or collected from waste of food industries and have become a benchmark in the study of biosorption. Moreover, it is an excellent model microorganism in biological research and its biomass is also considered as safe [18-20]. Therefore, biosorbents made from S. cerevisiae can be easily accepted by the public when applied especially to investigate the interactions of metal–microbe at molecular level [21]. The use of yeasts in model systems is particularly attractive because of the easy genetic manipulation and the availability of the complete genomic sequence knowledge on the molecular biology of the yeast is helpful to identify the molecular mechanism of biosorption in metal ion removal [22-24]. Transformation technique is an important tool in which exogenous DNA is introduced into a cell, resulting in genetic modification. To improve the biosorption capacity of the potential yeast biosorbent, a short metal-binding can be displayed in the cell’s surface with purpose to increase its heavy metal biosorption capacity from solutions. Among the heavy metal-binding in the cells, the phytochelatins (PCs) is one of the best studied so far [25]. The EC20 gene encodes a synthetic phytochelatin [25-27]. These peptides bind to metal ions, keeping them on their cell surface [28,29]. To verify the capacity of binding heavy metal on the fungal cell, progress has been made in exploring the cell surface ultra-structure of yeasts owing to the development of techniques such as the transmission electron microscopy with energydispersive spectrum analysis (TEM–EDS) [18]. |
| The aim of this study was to modify by genetic transformation a strain of S. cerevisiae to improve the biosorption of toxic heavy metals (Lead and Cadmium) and to compare the strain’s capacity of retaining heavy metals before the genetic transformation and after that. |
| Materials and Methods |
| Saccharomyces cerevisiae and Escherichia coli strains and growth media |
| S. cerevisiae W303-1A (MATa ade2-1 trp1-1 his3-1,15 leu2-3,112 ura3-1 ρ+canR) strain (R. Rothstein-Columbia University) was cultivated on rich glucose media (YPD - 1% yeast extract, 2% peptone, 2% dextrose) and minimal glucose media (0,67% yeast nitrogen bases, 2% glucose) supplemented with the appropriate auxotrophic supplements. The E. coli strain RR1 (Δ(gpt-proA)62, leuB6, thi-1, lacY1, hsdB20, rpsL20 (Strr), ara-14, galK2, xyl-5, mtl-1, supE44, mcrBB) (3183) was cultivated on LB plate (Luria Bertani) supplemented or not with ampicilin for plasmid selection. |
| EC20 synthesis, vectors construction and yeast transformation |
| EC20 was assembled from the oligonucleotides pairs EC20-1 and EC20-2 [30-32]. The complementary oligonucleotides were heated at 90oC for 2 min and hybridized, first at 55oC for 30 min followed by 37oC for 30 min. The double stranded fragment was digested, respectively, with BamHI and HindIII and inserted into YIp351 [33] and YCp22 containing the TRP1 marker [34] previously cut with the same restriction enzymes. YIp351-EC20 and YCp22-EC20 recombinant plasmids transformed W303-1A yeast strain following the procedure of Schiestl and Gietz [35]. YIp351-EC20 was linearized with BstXI and integrated into chromosomal LEU2 locus [30]. |
| Recombinant DNA manipulation |
| Standard methods were used for recombinant DNA manipulations, E. coli growth and transformation [36]. The recombinant plasmids YIp351-EC20 and YCp22-EC20 were isolated from E. coli in small and medium scale procedures [37,38]. For plasmid sequencing was used the Pure Yeild TM Plasmid Mini prep System kit (Promega Co., Madison, WI, USA). The obtained sequences were analyzed using Bio Edit program. |
| Metal ion solutions and biomass preparation |
| Two kinds of metal ion solution were used: solutions containing lead and cadmium. Metal ion solutions were made by dissolving analytical Pb(CH3COO)2. 3H20 (1g/L) and, CdCl2. 2,5H2O (0,5g/L) in deionized water. The fungal biomass was prepared in Sabouraud broth (Sb) (Oxoid, England), and incubated at 25oC for 5 days, and shaked at 150 rpm. After incubation, the pellets were harvested and washed with double-deionized water, corresponding to live biomass. The pH value of the solutions was adjusted. The conditions were optimized during the preliminary experiments to have all metals in a dissolved aqueous form and to achieve high biosorption capacity of all metal ions. |
| Transmission electronic microscopy |
| From the growth of fungi, broth was filtered to obtain biomass that was sent to the microtome sector to obtain the ultrathin sections. The ultrathin sections were examined by electron microscopy (JEOL- 1010) at IPEN and ICB department at the University of Sao Paulo. Transmission electron microscopy was performed with the strain of S. cerevisiae modified genetically and non-modified in this study with the purpose to reveal the presence of heavy metals in the fungal cell. |
| Results |
| Transformation of Saccharomyces cerevisiae |
| With regard to transformation of yeast, S. cerevisiae strain was transformed with YCp22-EC20 and YIp351-EC20 recombinant. YCp22-EC20 was chosen for yeast plasmid rescue and it was again sequenced confirming the stability of YCp22-EC20 in yeast. The transformed cells were tested for increased ability to chelate heavy metals into their cell surface, which showed an improvement in its capability to grow in medium amended with different lead and cadmium concentrations. |
| Transmission electronic microscopy |
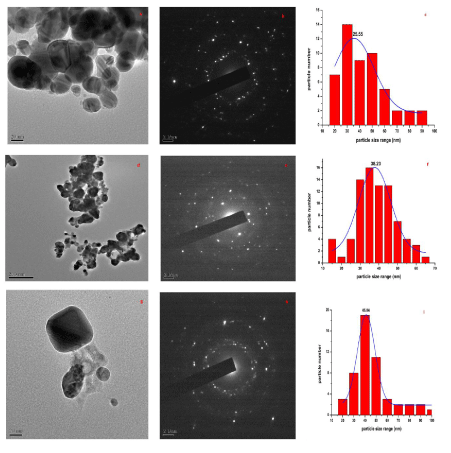
| Transmission electron microscopy images and EDS analysis of the cells of S. cerevisiae showed that heavy metals uptake have increased by transformed yeast cells and also revealed the presence of heavy metals mainly in the cell wall. Comparing to the control (Figure 1), the cells showed a high increase in their Pb2+ binding capacity (Figure 2a and b). The micrograph Figure 2a showed a transformed S. cerevisiae after genetic transformation and the lead accumulated in the wall cell. The micrograph Figure 2b showed a non-transformed S. cerevisiae and the wall cell with less lead accumulated. As indicated above, we observe that S. cerevisiae cells have a lower capacity of Cd2+ binding than Pb2+ binding (Figure 3a and 3b). TEM images and EDS analysis of S. cerevisiae grown in the presence of Cadmium after genetic transformation. The micrographs (Figure 2a) showed a transformed S. cerevisiae with cadmium accumulated in the wall cell. The micrograph (Figure 2b) showed a non-transformed S. cerevisiae and cell wall without cadmium accumulated. No heavy metals were detected in the control. As a result, during biosorption, the cells of S. cerevisiae released some substances without strongly damaging the cells integrity. The presence of electron-dense deposits was observed by transmission electron microscopy. |
| Discussion |
| Yeast cells have been used in the removal of heavy metal ions from wastewater [39-41]. S.cerevisiae could treat all major toxic heavy metals [1]. Metals are frequently internalized by the fungal cell and are accumulated into the vacuoles or can also be bound to low molecular weight proteins such as phytochelatins [41,42]. To improve metal uptake capacity, some techniques of biomass processing have been studied. Most biomass modification techniques described in the literature have shown to improve biosorption ability [43,44]. Metabolism-independent binding of metal ions in yeast cell is usually fast and large amounts of heavy metals can be bound to the cell [1]. In our study, S. cerevisiae cells were genetically transformed. Synthetic genes encoding for EC20 were synthesized as described previously [25,32]. According to our study, modified and non-modified strains were used to uptake lead and cadmium from broth. In studies of Bae [25], the EC20 gene was used to anchor EC20 on the surface of the cells, providing an increase in their heavy metal binding capacity. Furthermore, Biondo et al. [32] described the effects of the cell surface display of a synthetic phytochelatin in Cupriavidus metallidurans for enhanced metals bioremediation. Great progress has been made in exploring the cell surface ultra-structure of yeasts owing to the development of techniques such as the transmission electron microscopy with energy-dispersive spectrum analysis (TEM– EDS). In our experiments the majority of accumulated Pb2+ was found in the cell wall, indicating the involvement of functional groups of cell wall [45-47], in sequestering the Pb2+ ions. Moreover, Brady and Duncan [47] also found that biosorption can be achieved through complexation by cell wall and membrane hydroxylated components. Kunst and Roomans [48] found polyphosphate granules localized in or close to the cell vacuoles, which are the major storage vesicles for heavy metal ions in S. cerevisiae. Orlovich and Ashford [49] followed up on aforementioned studies and determined the exact vacuolar location for polyphosphate granules incorporation. Favero et al. [50] found that 80% of Cd2+ was bound to the hyphal cell walls in their study of Pleurotus ostreatus. In summary, the potential advantages in use of fungal biomass especially S. cerevisiae certainly could be applied for treatment of large scale heavy metals pollutants without the generation of toxic sludge. In the same context, genetic manipulation of S. cerevisiae could to increase the biosoption of heavy metals. |
| Acknowledgements |
| This work was supported by FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo). |
References
|
Figures at a glance
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 16472
- [From(publication date):
November-2015 - Jul 03, 2025] - Breakdown by view type
- HTML page views : 11505
- PDF downloads : 4967
