Plant Viruses
Received: 02-Oct-2023 / Manuscript No. acst-23-117571 / Editor assigned: 05-Oct-2023 / PreQC No. acst-23-117571 / Reviewed: 19-Oct-2023 / QC No. acst-23-117571 / Revised: 23-Oct-2023 / Manuscript No. acst-23-117571 / Published Date: 30-Oct-2023
Abstract
Viruses are mostly infectious microorganisms composed of a piece of nucleic acid and surrounding protein sometimes lipoproteins. They are parasites incapable of producing their own genetic materials important for living. All plants are affected by one or more viruses. Most have specific relationship with their host and vectors based on the nature of genetic material, their host and vectors bust some have wide range of hosts and vectors which makes them more infectious. The impact of viruses is increasing because of the increase in emerging disease and factors such as climate change, monocropping and material exchange are worsening the condition. But recent discoveries in molecular science revolutionized the knowledge and understanding of viruses. Even it contributing in exploiting beneficial aspects of viruses. Virous are very numerous and their rapid evolution complicated their classification. But viruses are classified based on morphology and the nature of their genetic material are mode of transmission are some of the criteria used for their classification. The interaction of viruses with their host quite complicated starting from their arrival to the release of their genetic martials inside the host. They use several mechanisms to identify and penetrate through surface of their host. Also, mechanism of their replication also varies where most the disassemble their protective coat progressively in order to avoid attack and undergoes post translation modification in order to diversify their infection and movement. The phenotype, biochemicals and their genetic materials are used for their identification but recent sciences such as multiplexing are making their identification easier. Integrated management of viruses effective for their control. Host factor produced by the plants attracts the vectors and avoiding the production of such chemicals is one of the resistance mechanisms used. Also, CRISPR aid RNAi are used for the development of resistance. The future is in the hand of this technologies and several other like virus-like plants (VLP) for identification, characterization and manipulation for diversified use.
Keywords
Virus; Vector; Host; Host factor; RNAi
Introduction
Viruses are very small microorganisms with infectious particles that are too small to be seen with simple light microscope. They are composed of small piece of nucleic acid which specifies two or more proteins surrounded by protein coat sometimes with lipoproteins. Viruses do not have a mechanism to produce and capture their own energy and they are not functionally active outside their host. They handle all the replication and translation using biochemical process of their host cell. Thus, viruses are obligate parasites in most cases disease causing because of this nature they are not regarded as true living organism [1,2]. Most organisms are the host for all viruses causing almost half of the reported emerging infectious diseases in plants. Conversely, some beneficial aspects of viruses have been discovered.
The history of virus begins in the late 19th century when it is first discovered at two different places in Dutch and Russia by Martuinus Beijerinck a microbiologist and Dmitrli Iwanowski a researcher. They described the virus as an unusual agent cause mosaic disease in tobacco leaf latter named Tobacco Mosaic Virus. Since then, the study of virus continues extensively and viruses known to cause a number of diseases by infecting all sorts of living organisms. The science that studies viruses known as Virology. It studies range of viruses from 16 to 2000nm which are generally organized inside the protein with the nucleic acids in cubic or helical symmetry shaped rod or isometric forms where rod shape is common [3 ,4 ].
Viruses have wide range of hosts and are known to infect all living organism. Most viruses are restricted to a particular or few hosts. Some infect bacteria called bacteriophage, others infect fungi known as mycovirus. There also viruses which infect algae, vertebrate, invertebrate and vascular plants. Viruses restricted to infect plants are plant viruses [5]. Plant viruses are important in the world agricultural production which known to reduce yield and quality resulting additional cost of several billions of dollars every year [6]. Globally, numerous plant viruses are presently known and almost all crop species are affected by one or more viruses. Recently, the highest impact is experienced with emerging disease because of their high incidence with large geographical range, rapid increase and pathogenicity. In addition to this, factors such as climate change, monocropping with high density and low genetic variability and increasing of in exchange of plant materials with trade and similar trends aggravated the situation. But the molecular techniques in the past few decades revolutionized the knowledge and understanding of viruses. Historically viruses have been perceived extremely a threat to living organisms including humans. These days, they are becoming beneficial in crop improvement, biomedicine and biotechnology industries [7 ].
There are numerous variants of viruses, this character of virus makes their classification and identification very resource full and difficult. There is no single classification system based their single property sufficient to identify and classify them. They are in continuous and rapid mutation changing their genetic makeup and new variants are continuously challenging. Numerous characters are used for the classification of viruses such as the disease they cause, the host they infect, mode of transmission, their vector and morphology. Moreover, recently sequence information becomes vital in the taxonomy and classification of viruses.
Presumably there are infectious RNA molecules more similar to virus later known to lack protein coding capacity which were classified under virus. These agents are called Viroids which also cause various economically important diseases. They typified by agent of potato spindle tuber disease [8]. They differ from virus in two ways where they lacked protein coat made only with naked RNA and do not produce any protein when they infect plant cell. There genome is smaller than virus and it is single strand and circular. Likewise, they do not produce their own protein and capable of use their host cell to produce their genetic material and move to other cell to infect the whole plant system.
Reviewed Topics
Theory of origin of virus
Theories on the evolution virus were hypothesized which are much debatable and there are three main theories. The first one says viruses are originated through progressive processes where genetic elements and pieces of genetic materials that are capable of moving within the genome gained ability to exit one cell and enter another. This theory is called progressive hypothesis and the second one is regressive hypothesis. It says viruses are evolved from more complex, possibly free-living organisms that lost genetic information overtime as they adopt parasitic approach to replication. The third one is opposite to the first hypothesis which states viruses predates or coevolved with the current cellular host [9 ]. The first two theories believe cell existed first but the third hypothesized that virus existed first. Due to this, some scientist argue that the third theories do not work since viruses are parasites of cells thus their origin should be after the exitance of cells [10 ].
Structure and composition
Viruses are composed of two main components that are the protective protein coat covering the inside structure and the genome made up of nucleic acid. In addition, in some viruses there is lipoprotein structure made of protein and lipids which used as an envelope protectant that protects the virus genome from the outside environment. Most viruses have genetic material composed of single strand RNA with positive sense like messenger RNA and some have negative polarity. But some of them have single or double strand DNA [11 ].
In viruses the protein shale called capsid arranged in one of the two forms of symmetry. The first one is helical which roughly elongated where the nucleic acid is arranged in highly ordered manner and the same helical conformation as proteinaceous capsids with two major variants of rigid rod and flexuous filaments. The other one is icosahedral which is nearly spherical with variants of bacilliform and twin virions composed of two incompletely joined icosahedra. In icosahedral virions the nucleic acid is partly ordered. In addition to their shape and structural forms, they are non-cellular multiply by assembling from pools of their structural components. They are dormant outside living cell, reproduce and become alive again inside the newly infected cells [12 ].
Taxonomy and classification
In the virus classification, different characteristics of the virus are used to group them in to families, genera and species. The characters used in the classification include morphology of the virus which can be represented by the identification of their shape and size in the electron microscope. Genome property is the other character used which include the size and the number of the genome, the similarity and relatedness of the genome and translation strategy. Biological and serological properties are also used in the classification. Biological properties related to the type host and their mode of transmission while serological properties related to the type of protein produced. The major classification property of all the character is the type of genetic material they have. It is depend on having single or double strand RNA or DNA [13,14]. Based on their genetic materials viruses classified in to five major groups.
Positive sense single strand RNA viruses (SSRNA+)
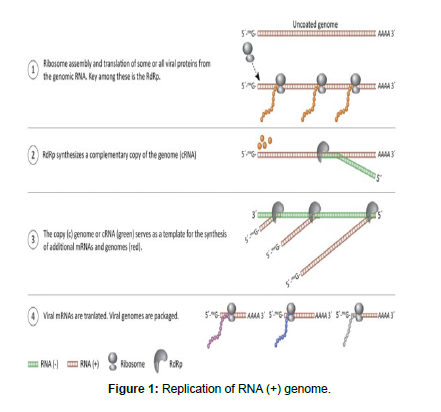
Positive strand RNA viruses have genomes that are functional mRNAs. Up on penetration into the host cell, ribosomes assemble on the genome to synthesize viral proteins. Genomes of positive strand RNA viruses are single-stranded molecules of RNA and may be capped and polyadenylated. During the replication cycle, among the first proteins to be synthesized are those needed to synthesize additional genomes and mRNAs. Thus, the infecting genome has two functions: It is an mRNA and also serves as the template for synthesis of additional viral RNAs. A functional definition of a positive-strand virus is that purified or chemically synthesized genomes are infectious [15 ].
Positive-strand RNA viruses often use large complexes of cellular membranes for genome replication. They actively modify host cell membranes to construct viral replication scaffolds. RdRp is a nonstructural protein, meaning that it is not found within the assembled virion. Instead, it is translated directly from the infecting genome shortly after penetration. RdRp and other viral proteins needed for viral RNA synthesis are encoded as a polyprotein that is cleaved by virally encoded proteases.
In the case of the picornaviruses and the flaviviruses, all viral proteins (structural and nonstructural) are synthesized as part of a single long polyprotein. Other positive-strand RNA viruses (i.e., togaviruses, coronaviruses, arteriviruses) synthesize an RdRp-containing polyprotein from genome-length mRNA, but use subgenomic mRNAs to encode structural and other proteins [16].
Negative sense single strand RNA viruses (SSRNA-)
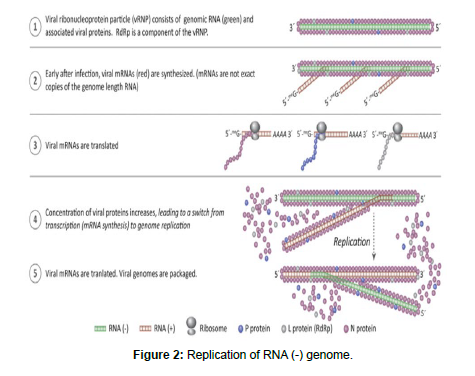
The closely related ambisense RNA viruses, and double-stranded RNA viruses. For each of these groups of viruses, the first synthetic event after genome penetration is transcription. This is accomplished by viral proteins (including viral RdRp) that enter cell with the genome. Transcription/replication complexes usually contain between two and four proteins (Figure 1).
They associate with the genome through interactions with RNAbinding nucleocapsid (N) or capsid proteins. Therefore, naked (purified away from protein) genomic RNA is not infectious, cannot be translated, and will eventually be degraded if transcription is blocked. Before genome replication can proceed, viral mRNAs must be transcribed and translated. Because the virion of a negative-strand RNA virus contains RdRp, it is possible to synthesize viral mRNAs in a test tube. If purified virions are gently lysed under appropriate buffer conditions, with the addition of NTPs, mRNAs will be transcribed in the test tube. However, genome RNA will not be synthesized under these conditions. Finally, the genomes of negative-strand RNA viruses are with the 3´ end to the left, opposite to the usual convention.
Negative-strand RNA viruses use the genome sense strand as the template for synthesis of all mRNAs. In contrast, viruses that use an ambisense coding strategy transcribe some mRNAs from the copy genome. There are virus families in which some members are considered negative-strand RNA viruses while others use an ambisense strategy.
Thus, these two strategies are closely related. Some ambisense viruses package copy genomes that can be used as templates for transcription, such that the full complement of viral genes can be transcribed soon after infection. It should be noted that packaged copy genomes are not mRNAs and are not translated (Figure 2).
Double strand RNA viruses
Family Reoviridae is a large family of dsRNA viruses. Reoviruses also have segmented genomes, packaging 11 to 12 segments of dsRNA. Reoviruses are nonenveloped and particles consist of two or three concentric icosahedral capsid layers. The genome segments are found within the innermost, T 5 1 icosahedral shell. A unique feature of the reovirus replication cycle is that the genome segments are transcribed from within the capsid. The mRNA products leave the capsid through pores at the vertices of the capsid [17 ].
Double strand DNA (RT) viruses
The Caulimoviridae is the only family because of their dsDNA genomes, might be effective gene vectors in plants. The family comprises six genera that form two groups’ caulimoviruses and badnaviruses these two groups differ in genome organization but have essentially the same replication methods. The replication does not involve integration into the host genome for transcription of the RNA but is from an episomal minichromosome. The template for replication is circular dsDNA and not the linear DNA with long terminal repeats characteristic of retroviruses. The DNA phase of the replication cycle is encapsidated rather than the RNA phase Thus, the Caulimoviridae are known as pararetroviruses. the replication cycle of pararetroviruses has two phases a nuclear phase where the viral DNA is transcribed by host DNA-dependent RNA polymerase cytoplasmic phase where the RNA product of transcription is reverse transcribed by virus-encoded RNA-dependent DNA polymerase or reverse transcriptase (RT) to give DNA (Figure 3).
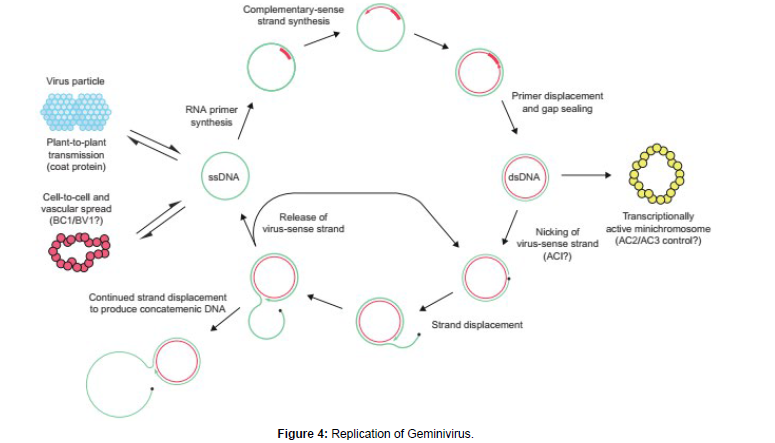
Single strand DNA viruses
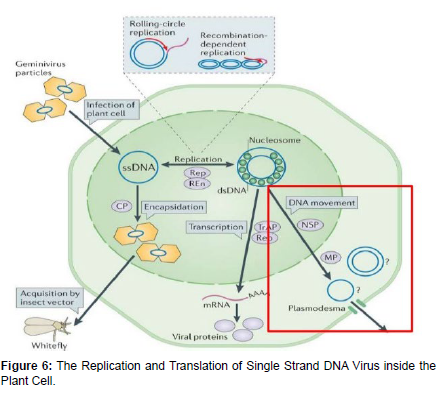
Two families Geminiviridae and Nanoviridae. There are four genera in the Geminiviridae mastreviruses, curtoviruses & topocuroviruses have monopartite genomes many of the begomoviruses have bipartite genomes. Persistent-non propagative: Geminivirus-diverse vector. DNA A contains all the information necessary for virus replication. DNA B involved in movement to the nucleus and between cells.
Persistent/non propagative: Phloem
Heat shock protein 70 (HSP70) and GroEL chaperone proteins have been implicated in transport to and stabilization in the hemolymph. RNAi Resistance (Tomato yellow leaf curl begmovirus). Replication is ssDNA>ssDNA via a dsDNA stage. Many of the features of replication of the two families are similar but there are some differences. Most is known about replication of the Geminiviridae depends upon many host functions. They replicate in differentiated cells that are in the G phase shut down most of their DNA replication activities. Thus, geminiviruses reactivate the replication activities that they require and convert the cell back to S phase [18 ].
Sign and symptoms
Symptoms are the effect of pathogen on growth and development plants and other organisms. There may be a change in body, function and disturbance in normal physiological processes. It is the result of sequence of events after the infection of the organism (Bos, 1977). Signs are physical evidence of a pathogen of plant and other organisms’ disease. In case of virus, observation of sign is very difficult and using symptom is more reliable. Viral diseases in the plants can be identified by different symptoms on the leaves, shoots, flower or fruits. But most could be confused with abiotic stresses, nutrient shortage and application of chemicals. Even symptoms could be shared between viruses. Although generalization is not possible and conclusive, there are symptoms that are distinctive.
The common effect of virus on the leaves includes narrowing, stunted growth, much reduced surface between veins, resembling damage of herbicide or hormone may be observed. Also leaves may be wrinkled, rolled, bent down or may show pale green or yellow pattern including spots, streaks, mottle, mosaic, vein clearing or banding pattern. Tuft stunted, yellow or brown streaks or spots on the steam could be observed. Also dwarfing of the steam is common. Flower may look small or distorted having white streaks without pigment. Fruit could be discolored and may show irregularly colored pattern such as marbling, ring spot and malformed contents [19,20]. In addition to external change, viruses produce internal change in reduction of chloroplast, necrosis of phloem and different kinds of inclusion body. Viral infection could be symptomatic or asymptomatic. Also, the infection could be acute or latent. Most of infection in the crop plants is acute where the symptoms will be visible with short time period. However, several plants could be infected latently without showing any symptoms. Although there are limited studies regarding latent infections, beneficial effects of latent infection are reported in number of studies and such infections might be common in nature than it is expected. They are beneficial in increasing the tolerance of some plants to other viruses or abiotic stress such as drought, temperature and salinity [21 ]. During the latency period the virus could remain dormant in the plant cell or become endogenous by integration of its genome sequence into the host genome. Then replication can follow. Indigenization of retrovirus is essential for an accomplishment of its life cycle but none of the reports indicated the integration viral DNA in plants required for replications [22] (Figure 4).
Mode of transmission and reproduction
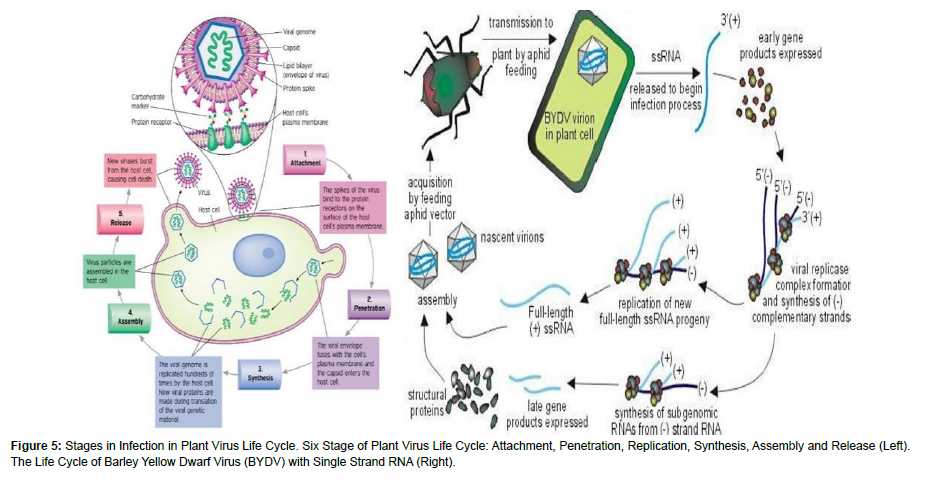
Plant viruses are obligate parasites starts their life cycle by penetration in to the host cell through different natural or wound openings resulted from different biotic and abiotic factors. Since the cell wall is strong enough to stop their penetration, viruses use some aids mostly vectors or introduced through wound made during cultural practices [23 ]. A limited number of viruses are transmitted through pollen to seed but infection could be accumulated in planting materials of vegetative propagated crops. They enter passively in to the cytoplasm through cuticle and cell wall that are damaged. Then they partially or completely remove their protein coat inside the cytoplasm. Then they transcribe their genome if they have DNA or directly go to translation if they have RNA genome. Translation will produce viral proteins which aids them to finish their life cycle but if their genetic material made of DNA, first they need to go to the nucleus for the process of transcription and the remining procedures [24 ]. In the process of translation viruses are needed to make at least three types proteins that used for nucleic acid, structural protein and capsid production. After replication process, the replicated protein combines with cellular protein to produce a complex protein that enable to manufactures multiple copies of the viral genome. These newly made genomes interact with structural proteins to form new virions [25].
There are three types of replication mechanisms. The first is making DNA directly from DNA and the second one is replication of RNA from RNA. The other is alternating between DNA and RNA. The virus that replicates DNA into DNA use the host machinery for this process but this is only active only during cell division. There is a mechanism to switch-on the host DNA replication enzyme in Gemini and nano viruses. Compared to the previous one DNA-RNA-DNA and RNARNA replications relatively unaffected by the plant cells. Caulimoviridae use DNA-RNA-DNA to replicate their genome which the first phase is effected by host enzyme while the second is virus coded phase. The RNA-RNA replication is most common in viral replication which mostly use its own coded enzymes and with some host coded factors. But why this mechanism is common is not known (Figure 5).
The next step in the virus reproduction cycle is movement of the virus into neighboring cells. Depending on the virus, the viral genomes or the virions are transported into neighboring cells through small channels called plasmodesmata that form connections between cells. Many viruses produce movement proteins that modify the plasmodesmata channels and facilitate viral movement into neighboring cells [26].
The process of cell-to-cell movement is relatively slow. It takes from a minute to a few hours for a virus to multiply in a cell and move to the next cell. To successfully colonize an entire plant, a virus needs to enter the vascular system of the plant. The process of systemic or long-distance transport normally precedes through the phloem sieve elements where viruses move passively with the flow of photosynthates. After quite rapid systemic spread of the virus in the phloem, the virus moves from the phloem into surrounding cells where it reproduces and spreads by cell-to-cell movement (Figure 6).
The time between initial infection of one or a few cells and systemic infection of the plant varies from a few days to a few weeks depending on the virus, host plant and environmental conditions. Transmission of the virus from one plant to another completes the virus life cycle [27 ].
Molecular plant virus interaction
The interaction of virus with the plant cell for the infection involves subsequent steps starting from arrival of virus particle in the plant tissue and entrance through different openings which is already discussed in the previous section. After arrival the following steps follows; penetration, uncoating, targeting, gene expression, gene replication, viral assembly (maturation) and finally release of new infectious virus. Also, viruses have antagonistic or synergetic relationship to each other during the course of double infection of host plant. Infection starts with viral particle disassembly, viral genome modification, cellular membrane modifications coupled with the formation of viral replication complex (VRC), viral genome encapsulation, cell to cell movement and finally long-distance transport. Since pre and post infection processes involve more of mechanical mechanism of interaction, let us see the molecular aspect of infection in more detail.
Viral infection starts with viral particle disassembly which is required for genome replication through the process of removing the protein coat of the virus called capsid. The early studies on tobacco mosaic virus (TMV) indicates virion de-coating process is passive triggered by the change in PH and positively cation concentration in the cell. The passive triggering process leading to the initiation of disassembly viral protein coat at 5’ end of encapsulated RNA. Some studies show the mechanism of disassembly through coupled bidirectional co-translational and co-replicational disassembly. Cotranslational mechanism involves limited degree of disassembly by protecting a part of RNA especially at the three ends from nuclease and translation of some parts specific around 5 ends. The exposeds portion of leader sequence forms an adjacent RNA binding site on the new protein surface directs the first stage of formation translation initiation complex from genomic RNA to translate 130K and 180K replicase proteins. This results the translation of two third of the coat protein subunits. The remaining one third at 3’ end is removed by virus replication complex (VRC) during minus strand RNA synthesis and the process is completed the removal coat protein molecules in 3’ to 5’ direction [28]. This kind of mechanism found to be 1.5 time more efficient than naked RNA infection of TMV. The processes of disassembly and replication (synthesis of negative strand RNA) takes place simultaneously after a few minutes of inoculation. Once started it continues rapidly and the synthesis of negative strand is more rapid than positive even positive strand is exposed before. The study by Wu & Shaw (1997) shows protein may only involve in the triggering the disassembly and the subsequent removal occurs by some other mechanism. The co-replicational mechanism only shown on TMV but the co-translational event is common in positive sense RNA viruses like pot viruses [29].
Viruses need pro-vital host factor that are suitable for viral infection which are vital cellular factors and resources necessary for viral infection and movement. They are essential for infection cycle and work in synchrony with viral factors. eIF4E and its isoforms are host factors recruited by plant pot viruses where viral VPg binds with them near the cap binding region which are essential for potyvirus infections. Also, in potato virus A (PVA) acidic ribosome protein P0 shown to promote infection by synergistically enhancing viral translation with VPg, eIF4E and its isoforms. In non-potyviruses Turnip yellow mosaic virus (TYMV), eEF1A interacts with the untranslated 3’ end to enhance viral genome translation [16]. But plant host factors also work against the establishment of the virus through the mechanisms such as autophagy, ubiquitination, mRNA decay and gene slicing of targeted viral component [30].
In viral replication, virus induced intercellular membrane structure provide a scaffold for tethering the VRC which confine viral replication process to specific safeguarded cytoplasmic site and deactivation of host antiviral mechanisms. Viral replication is coordinated by protein targeting and remodeling of the cellular membranes by viral proteins and co-opted host factors. Along with the remodeling of cellular membranes by viral proteins and the co-opted host factors. All VRC components must be summoned together at the replication site to form a functional unit for catalysis of viral replication. The VRC housed in the modified cellular membranes consists of viral replicase proteins, such as RNA dependent RNA polymerase (RdRp) and viral helicase, a viral RNA template and diverse host proteins, such as HSPs and RNA binding proteins (RBPs.
Viral proteins in the host plant may undergo post translational modification to increase functional diversity of infection with increasing protein diversity. After finalizing the translation and replication of their genetic materials viruses undergo long distance or cell to cell movement using the metabolic pathways through plasmodesmata (PD) which requires coordinated action of virus encoded movement proteins and host factors (Wang, 2015). In addition, genes in the host plant involved in the antiviral activities could be essential or non-essential based their effect on the host. There are essential gens like AGO1 which are active both in the antiviral activity and other additional function such as miRNA dependent regulation of gene expression and development. In contrast, genes like DCLl and DCL4 are non-essential that are redundant to each other which shows only mild leaf malformation (Garcia-Ruiz, 2019). When a virus infects the host cell, viral double stranded RNA (dsRNA) is synthesized by viral RNA dependent RNA polymerases (RdRp). From initially infected epidermal or mesophyll cell then move to the neighboring cells through plasmodia and systematically long distance through vascular tissues. During the infection of virus in plants the recognition of dsRNA of virus by the defense system the host plant led to RNA slicing by RNA interference (RNAi). To counteract antiviral activity, plant viruses encode RNA slicing suppressors. In this case, plant have evolved intercellular resistance (R) protein recognizing a pathogen encoded a virulence factor (Avr) leading to effector triggered immunity. Three scenarios have been proposed to explain super infection exclusion (SIE) in plants. The first is secondary virus enter in to the cell which is already infected. In this case, mechanisms like prevention of uncoating of secondary viruses, discharging of uncoated RNA and the negative strand may hybridize to the secondary viral RNA which ultimately results dicer mediated cleavage. The second scenario is the secondary virus enters cells that have been primed by Viral Small Interfering RNA (vsiRNA) but do not contain RNA of the primary virus. The last is the secondary virus enters cells distant from the primary infection. Phloem mobile long distant signal is sent to amplify the slicing response by endogenous RdRp which activates RISC and degrade RNA of secondary virus.
Vectors, alternative hosts and their spread
Viruses spread and transmitted from plant to plant with different agents. They could be transmitted vertically to from the plant to its progeny by vegetative propagation or horizontally from plant to plant though different agents. They can spread mechanically via pollen or biological agents called vectors like insects, nematodes and fungi. Virus infected weed, nearby plants, planting materials and remaining debris from the previous growing season also create favorable conditions for spread. In addition, large scale monocropping, transport of plant materials, increasing use of clonal planting materials and cultivation of virus susceptible plants aggravates the condition. But insect vectors take the line share of the transmission.
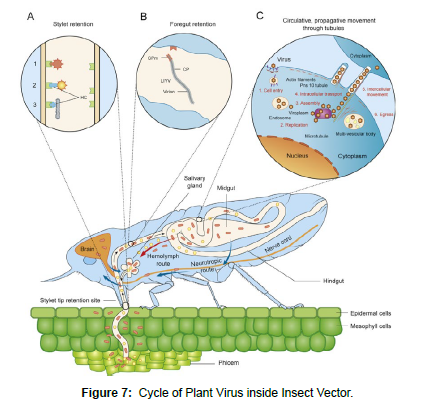
Agents called vectors actively transmit viruses from infected plant to healthy one during their feeding process. Arthropods, nematodes and parasitic fungi are the major vectors for transmission of plant viruses. Among those aphid and whiteflies are able to transmit the largest number of virus species. But transmission is very specific, each particular virus transmitted by only specific vector. Although fungi and nematodes are soil inhabiting and have little mobility, they have effective ways of persisting at specific site and make the transmission effective (Figure 7).
Viruses do not stay in the environment for longer time, only few of them like Tobacco Mosaic Virus (TMV) survive longer up to decades in the environment or on passive plant to plant mechanical transmission. Most virus actively transmitted with in short time period (Figure 8).
The interaction of virus and the vectors are specific mainly related to transmission specific to the host. The transmission is the end result of an attachment which can be rapidly absorbed by the external surface of the vector mouth part attached with special protein and released in to the host plant during feeding. In such case the transmission takes few minutes where such kind of relationship called non persistent [6]. The other relationship is called persistent where the virus circulates inside the body of vector like aphid moving through the gut wall to the body cavity and then to the saliva of thes vector which takes a minimum of 12 hours before transmission. In some cases, there is even further intimate relationships. The virus multiplies inside their vector. For this, the good example could be Tomato Spotted Wilt Virus inside thrips.
Persistent and more intimate interactions can cause direct or plant mediated effects on the insect vector which could modify the life cycle and feeding behavior of the vector. The relationship is mutual which increase the efficiency of feeding of the vector and for the virus it directs the vector to feed on the healthy plant which helps to continue its life cycle [14]. The study by Moreno-Delafuente et al. (2013) on tomato patho-system showed virus presence within B. tabaci body directly influenced vector settling, probing and feeding behavior in a way that enhance transmission efficiency and spread. The interaction was mutually beneficial because the efficiency of feeding increased after the aquation of the virus. Similar study done by Carmo-Sousa et al. (2016) on cucurbits aphis born yellow virus identified the modification of aphid vector change in feeding behavior in favoring cucumber host which are non-infected. Such interactions have an important role in increasing transmission and epidemiology viral diseases. Moreover, viruses with areal vectors persistent relationship with perennial host ensure survival during unfavorable growth of the plant.
Non persistent relation of viruses is shown in the relationship between single strand RNA positive sense (SSRNA+) viruses and aphids. This virus also shows semi persistent and persistent non propagative interaction with aphids, whiteflies and leaf hoppers. This specific group of viruses which shows persistent relation with aphids are luteoviridae. Also, double strand DNA (dsDNA) shows non persistent interaction with aphids. Other groups of viruses show persistent interactions with aphids, plant hopper and leaf hoppers which most require specific vector for their spread. But single strand DNA viruses like Geminiviruses have wide range of vectors (Table 1).
| Taxonomic families | Virus genus | Representative virus | Vector | Proteins | Initial entry |
|---|---|---|---|---|---|
| Potyviridae | Potyvirus | Tobacco etch virus (TEV) | Aphid | CP, HC-Pro | Stylet |
| Bromoviridae | Cucumovirus | Cucumber Mosaic Virus (CMV) | Aphid | CP | Stylet |
| Caulimoviridae | Caulimovirus | Cauliflower Mosaic Virus (CaMV) | Aphid | CP, P2, P3 | Stylet, acrostyle |
| Closteroviridae | Crinivirus | Lettuce Infectious Yellow Virus (LIYV) | Whitefly | CPm | Foregut |
| Luteoviridae1 | Luteovirus | Barley Yellow Dwarf Virus (BYDV) | Aphid | CP-ATP | Midgut, hindgut |
| Geminiviridae1 | Begomovirus | Tomato yellow leaf curl virus (TYLV) | Whitefly | CP | Midgut, filter chamber |
| Bunyaviridae2 | Tospovirus | Tomato spotted wilt virus (TSWV) | Thrips | GN | Midgut |
| Reoviridae2 | Phytoreovirus | Rice dwarf virus (RDV) | Leafhopper | P2+ | Midgut, filter chamber |
| Rhabidoviridae2 | Nucleorhabidovirdae | Maize mosaic virus (MMV) | Plant hopper | G | Midgut |
Table 1: Vectors and mode of their transmission in families of plant viruses [21] i.e. 1: Persistent non propagative virus; 2: Persistent propagative virus.
Viruses have alternative hosts that can serve as a bridge in the plant to plant spread and transmission or in a condition where the plants are in stress which helps the virus to continue their life cycle other than the vector. Most of the alternate host are weeds and nearby plants to the main host and could also be other main cultivated plants that are not most favorable to the virus. Different surveys on weed specious found that hundreds of weeds like amaranths, datura and solanum nigrum that are prevalent and favors thrips, white flies and aphid reproduction. Among them, one of the studies found Amaranthus hydrides have particular importance because of its fast growth and having numbers of generations per year. Also, more number of Amaranthus, Sonchus and Raphanus specious are identified that are vectors for tobacco, common bean, ground nut, brassica, sweet potato and lettuce. Sun flower and marigold were also found a potential alternative host for thrips.
Diagnosis and detection methods
The combined properties of viruses are important and much reliable for diagnosis than looking for symptoms. Phenotyping the disease can provide limited information and similar symptoms could be arising with different viruses. Integration is better because more than one property or responses are used for identification. The properties which use for diagnosis includes pathogenicity in such method bioassays using indicator plants are used. A response of some like plant genera of Nicotiania and Chenpodium for viral infection is consistent and distinctive used as an indicator. The other is transmissibility because specificity of the vector identification the vector provides information on the type pf the virus. With the help of electron microscope, the architecture of virus particle and presence viral structure could be identified by seeing its shape and size. The other method is immunology procedure where the viral particle injected in to the living organism and based antibodies response the virus could be identified. Based on this response of living cell a method called ELISA (Enzyme Linked Immunosorbent Assay) were develop which is a cheap and widely used technique in agriculture. Beside these more advanced, accurate and expensive methods are used. PCR (Polymerase Chain Reaction) method is one of them which uses the viral nucleic acid to identify its unique sequence by amplification using specific primers. It is a very sensitive and has specific procedure for virus detection. Recently, procedures that do not require any previous knowledge of viral sequence with much more efficiency and precision are introduced like High Throughput Sequencing. Also, there is a procedure called multiplexing which enables to detect and identify various viruses at once (Rubio et al., 2020).
Control and management measures
The control of viruses is very challenging because they are living in wide range of organisms and their life cycle is linked to many of them. Single control method for management of viral diseases is mostly ineffective. Thus, combining different measures will be effective in managing the transmission, spread and damage of plantss. Using a combination of cultural management (infected plant incineration, control of weed and neighboring crops), use of chemicals (herbicides and insecticides for weeds and vectors), breeding for resistant cultivars (artificial and natural resistance) and international legislation (certification and quarantine) will bring more effective management. The specific identified measurers include using of clean planting materials like apical meristem, crop hygiene, heat treatment, biological enemies, removal of alternative hosts and vectors using herbicides and insecticides. Also, field and planting materials certification, quarantining and improvement of resistance using both conventional breeding and genetic engineering found to be effective.
In viral transmission the role of vectors such as insects, fungi, nematodes and alternative hosts are very important for recycling of its life cycle and survival. The removal such agents will have a great role in reducing the viral population and the prevalence of the disease. In this regard, development of insect pest resistance is one of the successful options against for Cassava Mosaic Virus. African white fly (B. tabaci) is the major pest for the spread of the virus and development of resistance against this pest resulted promising level of resistance in several lines of Uganda, West Africa and South America. Similarly, white fly resistance was also developed in tomato against Tomato Leaf Curl Virus and Tomato Mottle Virus. The other source of infection that induces the infestation of the plant is the plant itself by producing deceptive attractive volatile substances which attract the insect vector. Such case is observed in tobacco producing aphid attractive volatiles for transmission of Cucumber Mosaic Virus (CMV). The viral resistance could be developed by avoiding such attraction which makes the plant not to produce such volatile secondary metabolites. This was proven by deletion of gene responsible producing 2b protein using mRNA interference that counter-the defense and diminishes resistance. Alternative hosts can be physically removed or herbicides can be used to remove them. For the vectors biological enemies are one way of reducing their population. But also, insecticides, fungicides and nematicides could also be used. During the use such chemicals, the effect on the beneficial insects like natural enemies should be considered. But till now, there is no single chemical that can effectively remove plant viruses is not discovered.
The prevention of disease establishment in areas where disease is not occurred before is the main principle of exclusion or quarantine. The movement and exchange of materials from place to place is monitored and the plant materials moving will be analyzed in the laboratory and field conditions by taking samples from different tissues. Sanitary certification will be used for virus free materials and fields for each and specific viruses and diseases [3].
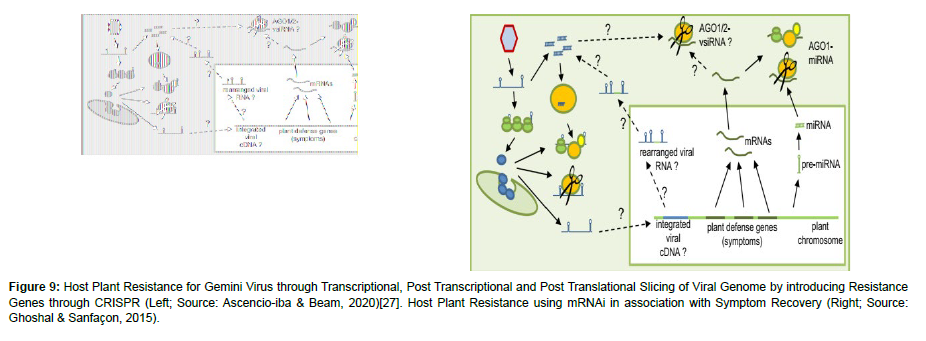
Immunization measures through introgression of resistant gene from cultivated or wild sources in improved cultivated crop using different techniques of breeding is highly effective source of resistance for viral diseases. The resistance gene could be introduced to the susceptible plant by introducing active resistant gene which is dominant. It recognizes the sequence of virus gene and results avirulence induced by cell death of the infected plant that prevent the virus from further movement to other cells. The other is introduction of passive resistance gene which is controlled by recessive alleles (Figure 9).
Figure 9: Host Plant Resistance for Gemini Virus through Transcriptional, Post Transcriptional and Post Translational Slicing of Viral Genome by introducing Resistance Genes through CRISPR (Left; Source: Ascencio-iba & Beam, 2020)[27]. Host Plant Resistance using mRNAi in association with Symptom Recovery (Right; Source: Ghoshal & Sanfaçon, 2015).
The dominant genes identified are R-genes belongs to nucleotide binding site leucine reach repeats (NBS-LRR) classes that specifically recognize viral avirulence (avr) gene product through the establishment of gene-for-gene interaction. An example for this could be Rx1 gene in potato that encode CC-NBS-LRR protein that mediate resistance to Potato potexvirus X (PVX) through recognition of PVX capsid. In recessive genes, resistance achieved through the absence host factor that are required to complete the cycle of the virus. Such resistances are used to develop partial and complete resistance against potyvirus in tomato and potato through the abundance of eIF4E resistance that make endogenous susceptible eIF4E inaccessible for its recruitment by viruses. An alternative strategy for introduction of new gene other than conventional breeding method is genetic engineering which identifies resistance from organisms within specious or inter specious and transfer the gene in the laboratory through different techniques including virus mediated gene transfer rather than biological hybridization (Nicaise, 2014). These days a more advanced gene transfer techniques which are capable of editing from a single base pair to a section of chromosome like gene editing are becoming popular. In this technique a method CRISPR found to be most effective which was first discovered in the bacterial immune system that recognize of viral chromosome.
Also, RNA mediated resistance called RNA silencing are used in combination of different techniques to confer host plant resistance. In this method, dsRNAs processed by ribonuclease type DICER (DCL) enzymes processed in to small RNAs (sRNAs) that are incorporated into RNA induced cytoplasmic silencing complex (RISC) which target mRNA to induce their cleavage. Natural resistance of Ty-1&3 genes against Tomato yellow leaf curl begomovirus (TYLCV) that encode RDR was observed that shows the tolerance of the plant.
Specifically, to each group of viruses there are different kind of resistance possible based on their vectors, structure and charters tics of the genome they have. RNA interference is known to be effective against most of the viruses while similar but different genes and slicers are used for each. In addition to RNAi recessive resistance (eIF4E) and hormone mediated resistance found to be effective against positive sense single strand RNA (SSRNA+) viruses. In double strand RNA (dsRNA) dominate and recessive resistance are found to be effective. Also, recessive resistances are used against double strand DNA (dsDNA).
Ecology and epidemiology
The study on concentration and interaction of plant viruses in the environment is called plant ecology. It deals with the factor influencing the behavior of a virus and interaction of viral population with complex host population in a variable environment. While the closely related science that studies the interaction and association of virus with its host plant and the disease is epidemiology. The study of epidemiology is within host population on cyclical development of disease in space and time. It also deals with the factors that influences the spread, determinants and distribution of viral diseases. It answers how and why the virus spread in the ecosystem. Thus, ecology and epidemiology studies help us to understand the complex interaction of viruses with their environment and host. They are essential to effectively control and predict viral diseases. These studies include different aspects of interaction with hosts like soil inhibiting vectors such as fungus and nematodes and their persistence at the site, vector intensity, seed transmission, virus survival during unfavorable plant growth. Also, in much improved understanding of the role climate in the activity and rate of virus spread, carrying capacity of habitat in the plant ecology, their integrated control and predictive model of virus epidemics.
Emerging viruses and viral diseases
These viruses are viruses that pose a major threat to plants especially to the food security which are rapidly spreading and occupy new areas that have not experienced the disease before. They could be new or discovered earlier and becoming apparent owing to the changing environment, ecosystem and emerging of new variants providing the opportunity to expand. The emergence and pandemics of viral diseases is continuously threatening the food security resulting a global economic impact of more than 300 billion USD annually.
Several reviews conducted form 1900 up until now indicate on several factors contributed for emergence of viral diseases. Some of the factors are rapid expansion of international trade, more frequent method of rapid transport, movement of plant away from form their domestication center, climate change and global warming which increased the vector population, exitance of multinational companies producing seed in one area and selling them in other and rapid mutation of viruses. Emergent viruses have two categories, one is the entire groups become emergent on global scale like white fly transmitted begomoviruses, trips transmitted tospoviruse and ciriniviruses. The other group is individual viruses which include viruses of tomato pepino mosaic virus, rice yellow mottle virus of the sobemovirus and potry virus infecting cucumber [31 ].
Conclusion
Viruses are small microbes composed of coat proteins and nucleic acid. They are obligate parasites use the host cell for their multiplication. They are classified based on their genetic composition DNA/RNA and the nature the genetic materials. Vectors and alternate hosts significantly contribute for thseir transmission. Acute/latent or persistent/non persistent relation host/vectors are existed between viruses and their vectors. Plants host factors and protein aid attraction and replication different disassembly coat proteins; replication and translation mechanisms coevolve with the host cell. Viruses diagnosed by phenotyping, bioassay, PCR and managed by Prophylactic or crop improvement measures. Although viruses wildly known by their negative impact on the production and quality of cultivated crops, new findings show they can be manipulated in genetic transfer and resistance breeding. The effect, attack and evolution of virus depend on their genetics, kind of relationship with their relation with vector and their host. Thus, development of resistance should consider all these through integrated management.
References
- Sastry SK, Mandal B, Hammond J, Scott SW, Briddon RW, et al. (2019) Encyclopedia of Plant Viruses and Viroids. New Delhi, India: Springer Nature.
- Gergerich RC, Dolja VV (2006) Introduction to Plant Viruses, the Invisible Foe. Plant Heal Instr 1-17.
- Rubio L, Galipienso L, Ferriol I (2020) Detection of Plant Viruses and Disease Management : Relevance of Genetic Diversity and Evolution Front. Plant Sci 11: 1-23.
- Mumford RA, Macarthur R, Boonham N (2016) The role and challenges of new diagnostic technology in plant biosecurity. Food Secur 8: 103-109.
- Takahashi H, Fukuhara T, Kitazawa H, Kormelink R (2019) Virus Latency and the Impact on Plants Front. Microbiolgy 10: 1-18.
- Dpv (2021) Descriptions of Plant Viruses 1-11.
- Wessner DR (2010) The Origins of Viruses. Nat. Eduation 9: 3-5.
- Nasir A, Romero-severson E, Claverie J (2020) Investigating the Concept and Origin of Viruses. Trends Microbiol 28: 959-967.
- Payne S (2017) Introduction to RNA Viruses. Viruses 97-105.
- R. I. Hamilton RI (2014) Replication of Plant Viruses. In Plant Virology, 5th Editio R Hull Ed Elsevier 5: 223-245.
- Dietzgen RG, Mann KS, Johnson KN (2016) Plant virus-insect vector interactions: Current and potential future research directions. Viruses 8: 1-21.
- Nicaise V (2014) Crop immunity against viruses : outcomes and future challenges. Front Plant Sci 5: 1-18.
- Bawden F (1945) Plant virus and virus diseases. Nature 155: 155-157.
- Jones RAC (2014) Plant virus ecology and epidemiology : Historical perspectives, recent progress and future prospects. Ann Appl Biol 164: 320-47.
- Syller J, Grupa A (2016) Antagonistic within-host interactions between plant viruses: Molecular basis and impact on viral and host fitness. Mol Plant Pathol 17: 769-782.
- Wang A (2015) Dissecting the Molecular Network of Virus-Plant Interactions: The Complex Roles of Host Factors. Annu Rev Phytopathol 53: 45-66.
- Wilson TMA (1984) Cotranslational disassembly of tobacco mosaic virus in vitro. Virology 137: 255-265.
- Wu X, Shaw JG (1997) Evidence that a viral replicase protein is involved in the disassembly of tobacco mosaic virus particles in vivo. Virology 239: 426-434.
- Garcia-Ruiz H (2019) Host factors against plant viruses. Mol Plant Pathol 20: 1588-1601.
- Jones RAC, Naidu RA (2019) Global Dimensions of Plant Virus Diseases : Current Status and Future Perspectives. Annu Rev Virol 20: 1-23.
- Whitfield AE, Falk BW, Rotenberg D (2015) Insect vector-mediated transmission of plant viruses. Virology 479-480: 278-289.
- Moreno-Delafuente A, Garzo E, Moreno A, Fereres A (2013) A Plant Virus Manipulates the Behavior of Its Whitefly Vector to Enhance Its Transmission Efficiency and Spread. PLoS One 8.
- Carmo-Sousa M, Moreno A, Plaza M, Garzo E, Fereres A, et al. (2016) Cucurbit aphid-borne yellows virus (CABYV) modifies the alighting, settling and probing behaviour of its vector Aphis gossypii favouring its own spread. Ann Appl Biol 169: 284-297.
- Aranda MA. Freitas-Asua J (2017) Ecology and diversity of plant viruses, and epidemiology of plant virus-induced diseases. Ann Appl Biol 171: 1-4.
- Anani J (2019) Physiological and Molecular Plant Pathology Evaluation of resistance to cassava mosaic disease in selected African cassava cultivars using combined molecular and greenhouse grafting tools ☆.Physiol Mol Plant Pathol 105: 47-53.
- Omongo CA (2012) African Cassava Whitefly, Bemisia tabaci, Resistance in African and South American Cassava Genotypes. J Integr Agric 11: 327-336.
- Ascencio-iba T, Beam K (2020) Geminivirus Resistance : A Minireview. Front Plant Sci 11: 1-9.
- Ghoshal B, Sanfaçon H (2015) Symptom recovery in virus-infected plants : Revisiting the role of RNA silencing mechanisms. Virology 479-480: 167-179.
- Rojas MR, Gilbertson RL (2008) Emerging Plant Viruses : a Diversity of Mechanisms and Opportunities. Plant Virus Evol 27-51.
- Jones RAC (2021) Global Plant Virus Disease Pandemics and Epidemics. Plants 233: 1-42.
- Goodin M, Verchot J (2021) Introduction to Special Issue of Plant Virus Emergence. Viruses 55: 13-15.
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Crossref , Indexed at
Citation: Siyoum YM, Girma BT (2023) Plant Viruses. Adv Crop Sci Tech 11: 629.
Copyright: © 2023 Siyoum YM, et al. This is an open-access article distributedunder the terms of the Creative Commons Attribution License, which permitsunrestricted use, distribution, and reproduction in any medium, provided theoriginal author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 2351
- [From(publication date): 0-2023 - Oct 21, 2025]
- Breakdown by view type
- HTML page views: 2026
- PDF downloads: 325