Research Article Open Access
Phylogenetic Characterization and Community Diversity of Hydrocarbon-Utilizing Bacteria in Soil Microcosms Enriched with AromaticHydrocarbons
| Ola Olapade A* | |
| Department of Biology and the Center for Sustainability and the Environment, Albion College, 611 East Porter Street, Albion, MI 49224, USA | |
| Corresponding Author : | Ola A. Olapade Department of Biology and the Center for Sustainability and the Environment Albion College, 611 East Porter Street Albion, MI 49224, USA Tel: (517) 629-0296 Fax: (517) 629-0264 E-mail: oolapade@albion.edu |
| Received July 01, 2015; Accepted July 08, 2015; Published July 10, 2015 | |
| Citation: Ola Olapade A (2015) Phylogenetic Characterization and Community Diversity of Hydrocarbon-Utilizing Bacteria in Soil Microcosms Enriched with Aromatic Hydrocarbons. J Bioremed Biodeg 6:305. doi:10.4172/2155-6199.1000305 | |
| Copyright: ©2015 Ola Olapade A. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
The phylogenetic identities and community diversity of indigenous bacterial populations in soil microcosms previously enriched with various mono-aromatic including benzene, ethyl-benzene and xylene (BEX) hydrocarbons were investigated using combinations of culture-based microbiological (phenotypic) as well as molecular (16S ribosomal RNA gene sequencing) approaches. A total of 45 bacterial isolates belonging to 5 distinct phyla were phylogenetically characterized among indigenous bacterial populations with putative hydrocarbon-degrading potentials in the soil microcosms. In general, bacterial members belonging to the γ-Proteobacteria (mostly species of Pseudomonas and Acinetobacter) were found to numerically dominate {representing between 60 to 94%} among the isolates from the three BEX-polluted microcosms. While, bacterial members belonging to the β-Proteobacteria (Comamonas and Delftia spp) and Firmicutes (Bacillus spp) were also represented. Results obtained from the community diversity calculations revealed relatively higher species richness in the benzene-spiked soils as compared to the other microcosms. Overall, the differences observed in bacterial phylotypes among the microcosms are probably attributable to the direct effects of the chemical properties of each hydrocarbon pollutant on the indigenous microbial community.
| Keywords |
| Bacterial isolates; 16S rRNA gene; Mono aromatic hydrocarbons; Pollution; Soils |
| Introduction |
| Aromatic hydrocarbons such as benzene, toluene, ethyl-benzene and xylene (i.e. BTEX) are ubiquitously found in various environments including soils and sediments where they occur naturally, mostly as part of lignin and as direct consequences of numerous anthropogenic practices [1-3]. These organic pollutants are known to often persist in such polluted environments partly due to their inherent toxicity and carcinogenic potentials as well as their hydrophobic properties which ensure their attachments to particulate organic matters in soil and sediments [4-7]. Therefore the widespread distribution of BTEX in various environments had resulted in several studies extensively exploring their potential biodegradation by indigenous microbial populations. However, while several indigenous bacterial species that are capable of degrading BTEX and other hydrocarbon pollutants have been successfully isolated from contaminated soils and sediments [8- 10], yet there is still currently paucity of information regarding the diversity and structural compositions of indigenous hydrocarbondegrading bacterial populations mostly responsible for in situ biodegradation of these common pollutants in various environments. |
| Previous studies have employed diverse approaches to elucidate hydrocarbon-contaminated environments under controlled microcosm and/or field conditions in order to delineate and implicate wide arrays of bacterial phyla capable of hydrocarbon utilization [11- 14]. For instance, Shi et al. [12] in their study utilized phylogenetic probes to examine in situ microbial community structures in regions of a contaminated, shallow sand aquifer that were also oxygen depleted. They concluded that fuel contamination at their study site fostered the proliferation of bacterial phylotypes that were previously minor constituents of the aquifer and were made up of diverse taxa that were dominated by members of the Proteobacteria. Similarly, Popp et al. [14] found bacterial members belonging to three subclasses (i.e. α-, β-, and γ-) of the Proteobacteria to dominate in two clone libraries that were generated by the analysis of small-subunit (SSU) rRNA genes from mineral-oil contaminated soils. In contrast, Greene et al. [13] observed successional and dynamic changes in the composition of soil microbial communities that were enriched with a mixture of aromatic hydrocarbons using reverse sample genome probing (RSGP), including various bacterial members belonging to the α-Proteobacteria (Sphingomonas spp), β- Proteobacteria (Alcaligenes spp) and γ-Proteobacteria (Pseudomonas spp), as well as the gram positive bacteria with high GC (Rhodococcus spp) and gram positive bacteria with low GC (Bacillus spp). |
| In this study, combinations of culture-based microbiological {i.e. phenotypic} and molecular {16S ribosomal RNA gene sequencing} approaches were used to isolate, characterize and identify 45 bacterial isolates with putative hydrocarbon-degrading capability in three soil microcosms previously spiked with BEX-hydrocarbons under controlled laboratory conditions. Additionally, various bioinformatics and analytical tools were employed for sequence alignments and diversity measurements. The main objective was to compare the bacterial community diversity in the hydrocarbon-degrading bacterial populations in response to the BEX hydrocarbon treatments between the microcosms. The goal was to determine whether, there will be similar or disparate hydrocarbon-utilizing bacterial populations among the indigenous microbial communities in the soil microcosms in response to the three separate BEX pollutants, based on their chemical properties. |
| Materials and Methods |
| Microcosm experiments |
| The soil samples used for the microcosm experiments in this study were collected within the premises of the Botanical Garden located in the Science Complex at Albion College in Albion, Michigan, USA (42.24°N 84.76°W). The collected soil was mostly silt and sand that was slightly greyish brown in coloration and made up of mostly fine particles. The soil pool collected was then divided into three equal subsamples (10 grams each) in sterile clay pots that were then used for the microcosm experiments. Briefly, the indigenous bacterial populations in the soils were exposed to the various BEX hydrocarbon substrates by adding and mixing thoroughly up to 10% V/W of each BEX substrate to soils in the sterile clay pots before incubating at 30°C in the dark for a period of about 2 weeks as previously described in Chikere et al. and Olapade et al. [15,16]. |
| Isolation of hydrocarbon-utilizing bacteria |
| After 2 weeks of incubation, 0.1 g of soil subsamples were removed from each treatment into sterile 10 mL Bushnell Haas (BH) broth (Difco & BBL, USA) that was supplemented with 0.025% of each of the hydrocarbon substrate. The tubes were then incubated with shaking at 180 rpm at 30°C in the dark for at least a minimum of 48 hours. After incubation, 0.2 mL was transferred from each broth onto sterile Bushnell Haas agar (BHA) plates (comprised per liter of: magnesium sulfate 0.2 g; calcium chloride 0.02 g; monopotassium phosphate 1.0 g; diammonium hydrogen phosphate 1.0 g; potassium nitrate 1.0 g and ferric chloride 0.05 g. 15 g of agar added to make the BH agar plates). Organic source was introduced through the vapor phase transfer by aspetically placing sterile Whatman filter papers previously impregnated {wet} with the respective hydrocarbon (BEX) substrate inside the lids of the Petri Plates, before incubating at 30°C in the dark for 48 hours. Bacterial isolates that grew on the BHA plates were then selected as putative hydrocarbon-degraders for subsequent screening and characterization according to standard methods [17]. All the isolates selected were screened for the 238 base pair long catechol 2, 3-dioxygenase [C230] gene fragment according to Mesarch et al. [18] using primers DEG-F (5’CGACCTGATC(AT)CGCA TGACCGA-3’ and DEG R (5’T(CT)AGGTCA(GT)(AC)ACGGTCA3’. All the hydrocarbon substrates utilized in this study were of analytical grade and obtained from Fisher Scientific. |
| Growth of bacterial isolates on hydrocarbon sources |
| Eleven bacterial isolates were selected based on their phylogenetic identities to represent the 11 main phyla obtained from the 45 total bacterial species in the three BEX microcosms. These selected bacterial isolates were then individually examined for their potential growth in the presence of various organic compounds as sole sources of carbon as previously described by Zhang et al. and Zhao et al. [9,10]. Briefly, 0.01% (W/V) of 12 different substrates selected because of their variable chemical compositions and ubiquity in contaminated environments, were added as sole carbon sources to previously sterilized BH medium and then incubated with shaking at 180 rpm at 30°C in the dark as done previously by Zhao et al. [10]. Solid substrates were either previously dissolved in 5% (W/V) diethyl ether before spraying onto the surface of the BH medium or placed directly on the plate lid in the case of naphthalene crystals [9]. Bacterial growth was then determined based on presence/absence of turbidity or the formation of clear zones around colonies on solid media |
| 16S ribosomal RNA gene amplification |
| 16S rRNA gene amplification was carried out by targeting the almost full-length 16S rRNA gene with the universal bacterial primer pair 27F (5’ AGA GGG AGA ACG CCT AGA TCG 3’) and 1492R (5’GGT TAC CTT GTT ACG ACT T3’) as previously described by Olapade [19]. Briefly, the PCR aliquots were heated at 95°C for 5 minutes following by 30 cycles of the following: 95°C for 5 minutes, 55°C for 1 minute and 72°C for 2 minutes. The amplified PCR products were then confirmed on agarose gel and purified with a QIA quick PCR purification kit (QIAGEN, Valencia, CA) before subsequently utilized for sequencing |
| Phylogenetic and community diversity analysis |
| The bacterial isolates from the soil microcosms that were successfully sequenced were analyzed using the Sequencher program (version 4.5; Gene Codes Co., Ann Arbor, MI) and then compared with previously published GenBank sequences using the BLAST system to determine their close relatives [20]. Sequence alignments and other manual editing were carried also out with ClustalW according to Perriere et al. [21]. Community diversity measurement for species richness, Shannon Index, and species evenness were also calculated among the isolates obtained from the three microcosms. Additionally, alpha, beta and gamma diversity calculations were carried out according to Whittaker [22]. |
| Nucleotide sequence accession numbers |
| The 45 nucleotide gene sequences obtained from this study have already been submitted to DDBJ/GenBank/EMBL under accession numbers LC018399 to LC018443. |
| Results |
| Hydrocarbon-utilizing bacterial isolation |
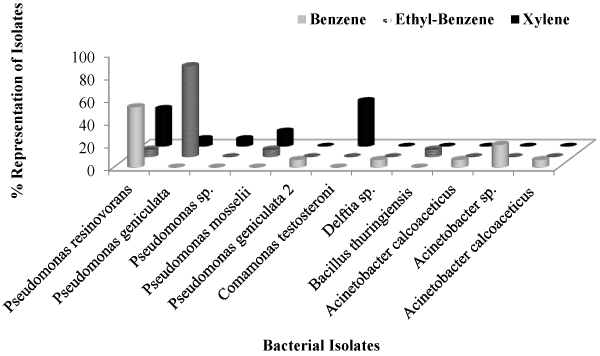
| Forty-five (45) bacterial isolates were successfully obtained from the hydrocarbon-contaminated soils on solid BHA media after exposure to benzene, ethyl-benzene and xylene under controlled laboratory conditions. The various phenotypic characteristics of these bacterial isolates are as listed in (Table 1). Briefly, the isolates are mostly gram negative, rod-shaped, endospore negative, but lipase and catalase positive, among other biochemical tests performed. Majority of the representative isolates were found to belong to the subclass of gamma Proteobacteria including several species of Pseudomonas and Acinetobacter (Tables 2 and 3). Comparatively, more bacterial isolates were obtained from the benzene-spiked soil microcosm (Figure 1) as indicated by a higher Shannon-Weiner diversity index of 13.07 as compared to relatively lower indices (8.35) and 9.45 in the ethylbenzene and xylene microcosms, respectively (Table 4). Also, results obtained from Alpha Diversity calculation revealed relatively higher species richness [6] in benzene-spiked soils to the lowest [4] in the ethyl-benzene microcosm. |
| Growth on carbon substrates |
| The potential of 11 of the representative bacterial isolates (i.e. 6 isolated from the Benzene, 2 from ethyl-benzene and 3 from the xylene microcosms) to grow in the presence of 12 different carbon sources were examined. The organic substrates including the BTEX compounds, methanol, naphthalene, and phenanthrene among others, were selected because of their composition as part of various petroleum products and their widespread distribution in contaminated soils (Table 5). All of the bacterial isolates examined were able to grow, with varying degree in the presence of benzene, ethyl-benzene, xylene, ethanol, naphthalene, and phenanthrene. While 9 out of the 11 bacterial isolates were able to utilize both benzene and phenol, however, none of the isolates appeared to show noticeable growth in both dimethylsulphoxide (DMSO) and salicylic acid (SA). |
| Discussion |
| The ecological importance of accurately delineating the bacterial composition and diversity in hydrocarbon-contaminated soil can never be overemphasized, especially based on previous studies that have documented significant shifts in bacterial community compositions in various environments during enrichments with various organic substrates [7,12-14,16,23]. In particular, Shi et al. [12] utilized 16S rRNA probing to explore both pristine and contaminated aquifers and found that toluene exposure significantly enhanced the proliferation of bacterial phylotypes that were previously minor constituents of the contaminated aquifer community examined. Their results suggested that toluene treatment probably helped in the identification and characterization of toluene-degrading phyla within the hydrocarbon utilizing assemblages. Also, another study corroborated this potential shift by utilizing 16S rRNA clone libraries to reveal that disparate bacterial phylotypes responded when exposed to toluene treatments in two separately contaminated soil microcosms that were then examined under controlled laboratory conditions [16]. |
| The Shannon-Weiner diversity indices obtained in the three microcosms i.e. 13.07 in the benzene spiked microcosm, 8.35 (ethylbenzene) and 9.45 (xylene) appeared to be relatively higher than those previously reported in similar studies that were based on examining microbial diversity in contaminated soils [14,23,24]. Comparatively, some of these studies documented lower Shannon-Weiner diversity indices that ranged from about 1.45 to upwards of 3.93 in their studied environments. However, it should be pointed out that, the relatively high diversity indices recorded in this present study could partly be attributable to the use of top soils with mostly biologically active compositions as compared to such soils with aged petroleum contamination [25] or those mostly made of sandy and stony soils from Antarctica [23]. |
| Furthermore, the results of both alpha and beta diversity measures probably suggest slight differences in the preferences of the indigenous hydrocarbon degraders for the three separate hydrocarbon substrates used in the BEX-spiked microcosms. Since previous studies have also indicated individual preferences for BTEX by microbial consortia under controlled conditions typically in the order of toluene-benzeneethylbenze- and xylene [26]. Therefore it can be suggested that the differences observed in bacterial diversity among the microcosms are partly attributable to the chemical properties of the hydrocarbon substrates. Comparatively, utilization of benzene as a substrate has been found to be most widely preferred by soil microbial communities followed closely by preference for toluene, the xylenes, styrene and naphthalene in that order [13]. |
| The numerical dominance of bacterial members belonging to the γ- Proteobacteria, mainly species of Pseudomonas and Acinetobacter, in this study appeared to further corroborate previous documentations that have also revealed high occurrences or community shift towards these particular bacterial phylotype, among indigenous hydrocarbondegrading microbial communities in response to hydrocarbon contamination [16,27,28]. Specifically, members of the pseudomonads observed to be dominant in the three microcosms examined in this study have also been shown to be prolific hydrocarbon degraders in several studies of contaminated environments [9,15,28-30]. Probably because the pseudomonads are copiotrophic with propensity for high concentrations of nutrients [31], thereby giving them the edge in typically outnumbering other bacterial competitors within any contaminated environment. The Acinetobacter species, which were also well represented within the BEX-polluted soil microcosms examined, have been previously shown to have the potentials of producing various biosurfactants that can be used to solubilize and biodegrade hydrocarbon compounds [32]. Overall, the differences observed in the bacterial phylotypes isolated from the three polluted soil microcosms in this study, suggests that the shifts in community compositions are probably attributable to variations in the chemical properties of the BEX pollutants on the indigenous hydrocarbon-degrading bacterial assemblages within the soil assemblages |
| Conclusion |
| This study has documented changes in hydrocarbon-degrading bacterial assemblages within the same soil community in response to varying hydrocarbon substrates. The observation further corroborates previous reports of such shifts in indigenous microbial community structures and diversity, mostly towards the dominance of taxa with hydrocarbon-degrading capabilities. Potential shifts in microbial community compositions in polluted environments are partly attributable to the metabolic proficiency of the assemblages as well as the type and complexity of the hydrocarbon pollutants. |
| Acknowledgements |
| Sincere thanks to my students, colleagues and staff members in the Science Complex and the Provost’s Office at Albion College for various support during the study period. |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 | Table 4 | Table 5 |
Figures at a glance
 |
| Figure 1 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 14021
- [From(publication date):
July-2015 - Apr 04, 2025] - Breakdown by view type
- HTML page views : 9436
- PDF downloads : 4585
