Research Article Open Access
Phenotypic Diversity Studies on Selected Kenyan and Tanzanian Rice (Oryza sativa L) Genotypes Based on Grain and Kernel Traits
| Mawia Musyoki A1*, Wambua Kioko F1, Agyirifo Daniel1, Nyamai Wavinya D1, Matheri Felix1, Langat Chemtai R1, Njagi Mwenda S1, Arika Arika W1, Gaichu Muthee D1, Ngari Ngithi L1, Ngugi Piero M1 and Karau Muriira G2 | |
| 1Department of Biochemistry and Biotechnology, School of Pure and Applied Sciences, Kenyatta University, P.O. Box 43844-00100, Nairobi, Kenya | |
| 2Molecular Biology Laboratory, Kenya Bureau of Standards, Nairobi, Kenya | |
| Corresponding Author : | Mawia Musyoki A Department of Biochemistry and Biotechnology School of Pure and Applied Sciences Kenyatta University, Nairobi, Kenya Tel: +254 711 953 234 E-mail: amosyokis@gmail.com |
| Received: September 19, 2015; Accepted: October 01, 2015; Published: October 03, 2015 | |
| Citation: Mawia Musyoki A, Wambua Kioko F, Daniel A, Nyamai Wavinya D, Felix M, et al. (2015) Phenotypic Diversity Studies on Selected Kenyan and Tanzanian Rice (Oryza sativa L) Genotypes Based on Grain and Kernel Traits. J Rice Res 3:150. doi:10.4172/2375-4338.1000150 | |
| Copyright: © 2015 Mawia Musyoki A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
| Related article at Pubmed, Scholar Google | |
Visit for more related articles at Rice Research: Open Access
Abstract
Phenotypic characterization of rice (Oryza sativa L.) provides useful information regarding preservation of diversity and selection of parental genotypes with superior traits in plant breeding program. The main objective of the present study was to characterize 13 selected rice genotypes from Kenya and Tanzania based on 7 grain and kernel traits. Minitab 15.0 software was used to analyze data. The 7 traits showed highly significant differences among the improved and local landraces. A dendrogram was constructed from data set of mean values of grain and kernel traits and showed two super clusters; I and II. Principal component analysis revealed that all the seven quantitative traits significantly influenced the variation in these genotypes.
| Keywords |
| Oryza sativa L; Dendrogram; Principal component analysis |
| Introduction |
| Rice (Oryza spices) is a monocotyledonous plant belonging to the family Granineae and subfamily Oryzoidea. It is ranked second to wheat among the most cultivated cereals in the world. Due to its importance as a food crop, rice is being planted on approximately 11% of the Earth’s cultivated land area. During crop improvement strategies, selection on breeding lines depends on a given set of criteria found suitable to a particular environment and for specific application. This process has led to development of morphologically related genotypes. Phenotypic similarity poses threats of epidemic of pests and diseases. To address this problem, phenotypic characterization is important in breeding program to avoid this inherent danger of phenotypic uniformity. In addition, landraces offers valuable genetic materials that can be utilized in future crop development and improvement programs. High yielding varieties which are the back bone of green revolution have led to erosion of landraces and wild varieties of rice5. Importance of landraces can never be ignored in agriculture system. This is because improvement in existing varieties depends upon desirable genes which are possibly present in landraces and wild varieties only. Therefore, characterization of phenotypic diversity on existing landraces of rice reveals important traits of interest that can be utilized in rice improvement programs. A number of research studies on phenotypic diversity assessment of various rice varieties around the world based on grain and kernel traits have been carried out. However, phenotypic diversity studies on rice genotypes from Kenya and Tanzania based on grain and kernel traits has not yet been studied before. Therefore, the objectives of this study were to determine the phenotypic diversity on selected rice (Oryza sativa L) genotypes from Kenya and Tanzania along other 2 genotypes from Philippine based on 7 grain and kernel traits and to identify the traits that contribute to the total variation among the rice genotypes studied. Information generated from phenotyping these genotypes can be utilized in rice breeding programs [1-6]. |
| Materials and Methods |
| Plant materials |
| A total of 13 rice genotypes comprising of local landraces and improved rice genotypes were collected from Mwea Irrigation Agricultural Development Centre (MIAD) in Mwea, Kenya and Kilimanjaro Agricultural Research Institute in Moshi, Tanzania. The name, country of origin and category of the rice genotype chosen for the study are given in (Table 1). |
| Measurement of grains and kernel traits |
| The following seven (7) phenotypic traits were measured in this study; grain length (GL), grain breadth (GB), grain length/breadth (G-L/B), kernel length (KL), kernel breadth (KB), kernel length/ breadth (K-L/B) and 100 grain weight (100 GW). For each of the rice genotype, 10 grains were randomly selected and their measurement taken using a digital vernier caliper. Weight of 100 rice grains from each genotype was determined using an electronic weighing balance (Mettler toledo), and average weight ± standard error mean recorded. |
| Data management and analysis |
| To determine phenotypic relatedness based on grain and kernel traits of the 13 rice genotypes, the data was analyzed statistically for the difference in means for the 7 grain and kernel traits measurement through ANOVA followed by Tukey’s post hoc to separate means. Cluster analysis yielded a dendrogram that was used to examine the phenotypic relatedness among the 13 rice genotypes. To assess the underlying source of variation in morphology based on the 7 grain and kernel traits among the 13 rice genotypes, Principal component analysis (PCA) was carried out. All analyses were done using software Minitab 15.0 (State College Pennsylvania-USA). |
| Results |
| Measurement of grain and kernel traits |
| The measurement of the grain and kernel traits for the 13 rice genotypes and their mean values are shown in Table 2. From the table, it can be seen that Supa, a local landrace rice genotype from Tanzania had the highest value of the grain length (10.48) while ITA 310, an improved rice genotype from Kenya had the lowest value of the grain length (9.00). In terms of grain breadth, Supa, a local landrace rice genotype from Tanzania had the highest value of the grain breadth (2.14) while ITA 310, an improved rice genotype from Kenya had the lowest value of grain breadth (1.79). Furthermore, it was observed that Wahiwahi, a local landrace rice genotype from Tanzania reported the highest value of grain length/breadth ratio (5.26) whereas IR 64, an improved rice genotype from Philippine had the lowest value of grain length/breadth ratio (4.60). Moreover, Supa, a local landrace rice genotype from Tanzania had the highest value of the kernel length (7.78) while ITA 310, an improved genotype from Kenya recorded the lowest value of kernel length (6.54). In terms of kernel breadth, Supa, a local landrace rice genotype from Tanzania had the highest value of the kernel breadth (1.90) while ITA 310, an improved genotype from Kenya recorded the lowest value of kernel breadth (1.60). In terms of kernel length/breadth ratio, Kilombero, a local landrace rice genotype from Tanzania had the highest value of the kernel length/breadth ratio (4.45) while BW 196, an improved genotype from Kenya had the lowest value of the kernel length/breadth ratio (3.68). In terms of grain weight, Supa, a local landrace rice genotype from Tanzania had the highest value of the grain weight (2.91) while ITA 310, an improved genotype from Kenya recorded the lowest value of grain weight (1.67) (Table 2). |
| Cluster analysis |
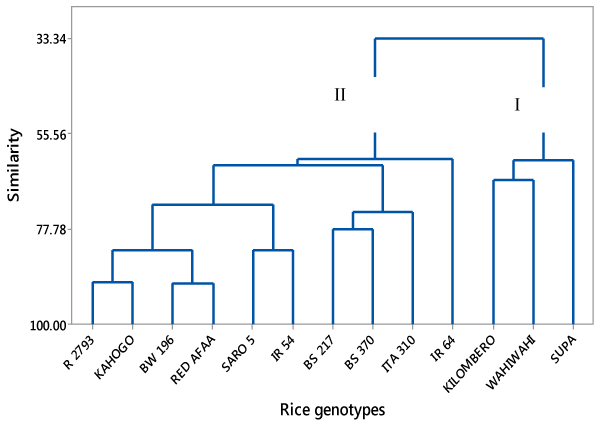
| A dendrogram was constructed from data set of mean values of the 7 grain and kernel traits and showed two super clusters; I and II as shown in (Fig 1). Super cluster I comprised of Kilombero, Wahiwahi and Supa genotypes both indigenous rice genotypes from Tanzania. On the other hand, super cluster II comprised of 10 rice genotypes; 5 improved rice genotypes from Kenya, 2 improved rice genotypes from Philippine, 1 improved rice genotype from Tanzania and 2 indigenous rice genotypes from Tanzania. Super cluster II was more diverse and could further be divided into 2 sub groups. |
| Principal component analysis (PCA) |
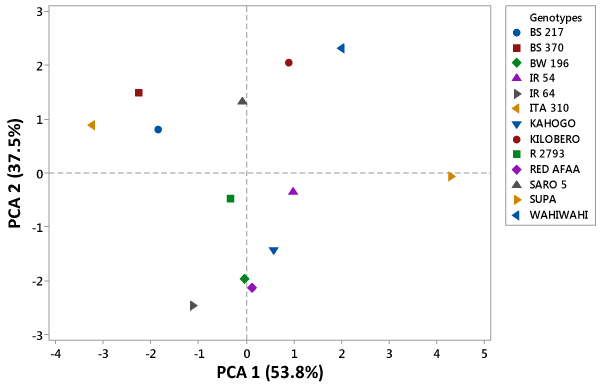
| The PCA was performed for all the 7 grain and kernel traits among the 13 rice genotypes as indicated in table. Out of seven, two principal components exhibited more than one Eigen value and showed about 91.3% variability among the traits studied. The PC1 had 53.8%, PC2 showed 37.5% and PC3 exhibited 5.4% variability among the genotypes for the traits under study. Principal component one (PC1), principal component two (PC2) and principal component three (PC3) had Eigen values of 3.77, 2.62, 0.38 and 0.14 respectively. The first PC was highly and positively correlated to grain length, grain breadth, kernel length and 100 grain weight. However, it was negatively correlated to grain length/breadth ratio. In the second PC, grain length/breadth ratio, kernel length and kernel length/breadth ratio were highly and positively correlated traits. On the other hand, it was highly and negatively correlated to grain breadth and kernel breadth. The third PC exhibited high positive correlation between kernel length and kernel length/breadth ratio. However, it was highly and negatively correlated to grain length and grain length/breadth ratio as shown in fig. Based on the 7 grain and kernel traits, the 13 rice genotypes were clustered into four groups as shown in figure 1. The first quadrant was made up of Kilombero and Wahiwahi. The second quadrant grouped Saro 5, BS 217, ITA 310 and BS 370 together. R 2793, IR 64 and BW 196 were grouped into third quadrant. The forth quadrant consisted of Red Afaa, Kahogo, IR 54 and Supa genotypes (Table 3 and Figure 2). |
| Discussion |
| Diversity analyses of germplasm collections of several crop species have revealed considerable variability for a wide range of traits. The wide range of diversity in phenotypic traits have proved a useful tool in classification of plants and the information obtained could be of high interest to plant breeders in development of plant species with desirable agronomic and nutritional qualities [7,8]. From the ANOVA table, it was evident that the means of the measured traits varied significantly among the 13 rice genotypes included in this study. The differences in mean values of grain length across the 13 rice genotypes studied could be as a result of different expression levels of GS3 gene that confers grain length in rice. In addition, the differences in mean values of grain breadth and grain weight across the 13 rice genotypes studied could be as a result of different expression levels of GW2 gene within the rice genome. Reduced expression of GW2 gene increases grain breadth, resulting in enhanced grain weight, whereas over expression decreases grain breadth and weight. Moreover, the differences in mean values of kernel length across the 13 rice genotypes studied could be as a result of different expression levels of Kne gene that confers kernel length in rice. These results were consistent with findings of. Genotypes that were clustered together indicated that they shared similar traits. For example, three improved rice genotypes namely BS 370, BS 217 and ITA 310 both from Kenya were grouped together and this showed that they have similar grain and kernel traits. Genetic improvement of these genotypes could have resulted in sharing of similar genes that influenced their phenotypic characteristics. On the other hand, genotype R 2793 and BW 196 both improved genotypes from Kenya formed separate clusters implying that they are phenotypically distinct from each other as far as grain and kernel traits are concerned [9-12]. |
| Principal component analysis measures the importance and contribution of each component to the total variance while each coefficient of proper vectors indicates the degree of contribution of every original variable with which each principal component is associated. In PC1, the traits that accounted for most of the 53.8% observed variability among 13 genotypes included grain length, grain breadth, kernel length, kernel breadth and 100 grain weight. Therefore, this implied that these traits contributed most to the diversity captured in the first principal component. Grain length/breadth ratio, kernel length and kernel length/breadth ratio were the most important traits that highly contributed to variation patterns among the genotypes studied in PC2. Moreover, in PC3 only 4 traits contributed highly to variation patterns among the genotypes studied. They included: grain length, grain length/breadth ratio, kernel length and kernel length/ breadth ratio. Kernel length contributed to variation across all the three principal components indicating that is one of the most important agronomic traits in rice. These results agree with the findings of. The phenotypic diversity witnessed among the rice genotypes studied could be used as possible parents for rice improvement program [13,14]. |
| Conclusion |
| Phenotyping of germplasm materials is an important undertaking in genetic resource conservation to ensure efficient conservation management as well as its effective utilization especially in breeding programs. In this study, 13 rice genotypes were characterized to assess their phenotypic diversity and showed considerable variability based on the 7 grain and kernel traits. There was a moderate significant difference in mean values of the measured traits. Out of the 7 traits measured, Supa had the highest number of 5 traits with highest mean values, that is, grain length, grain breadth, kernel length, kernel breadth and grain weight while ITA 310 had the highest number of 5 traits with lowest mean values, that is, grain length, grain breadth, kernel length, kernel breadth and grain weight. Cluster analysis based on the 7 grain and kernel traits clustered the 13 rice genotypes into 2 major groups. Cluster I was less diverse since it consisted of only 3 genotypes while cluster II was more diverse and consisted of 10 genotypes. Such groupings are useful to breeders in identifying possible genotypes that may be used as parents in breeding for any of the phenotypic traits that were studied. The PCA revealed that all the traits contributed significantly to variation patterns among the genotypes studied. Above all, the information generated will reduce the overall time required by rice breeders to screen large populations for potential breeding stock. |
References |
|
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 |
Figures at a glance
 |
 |
| Figure 1 | Figure 2 |
Relevant Topics
- Basmati Rice
- Drought Tolerence
- Golden Rice
- Leaf Diseases
- Long Grain Rice
- Par Boiled Rice
- Raw Rice
- Rice
- Rice and Aquaculture
- Rice and Nutrition
- Rice Blast
- Rice Bran
- Rice Diseases
- Rice Economics
- Rice Genome
- Rice husk
- Rice production
- Rice research
- Rice Yield
- Sticky Rice
- Stress Resistant Rice
- Unpolished Rice
- White Rice
Recommended Journals
Article Tools
Article Usage
- Total views: 14622
- [From(publication date):
November-2015 - Jul 06, 2025] - Breakdown by view type
- HTML page views : 9849
- PDF downloads : 4773
