Performance of Laboratory Methods for Detection of Polymyxin-Resistance in Gram-Negative Bacillus
DOI: 10.4172/2161-0681.1000001
Abstract
Resistance to polymyxin, mediated by chromosomal mutations and plasmid-borne mcr genes, is increasingly being reported not only in clinical bacteria but also in animals, farms, foods, and the environment. In this study, we evaluated a test capable of detecting resistance to polymyxins during antimicrobial susceptibility testing by the agar dilution test. We evaluated 521 consecutive isolates from A. baumannii, P. aeruginosa and Enterobacteriaceae and 57 polymyxin-resistant K. pneumoniae. We performed tests by the agar dilution method for colistin and polymyxin B (colispot and polyspot, respectively), the Broth Micro Dilution (BMD) method, and the VITEK ® 2 automated system for colistin. The colispot and polyspot tests were evaluated in concentrations of 2.0, 4.0 and 8.0 μg/mL. Considering BMD as a reference method, 420 isolates were shown to be colistin sensitive, 158 isolates were colistin resistant, 423 isolates were sensitive to polymyxin B and 155 isolates were resistant to polymyxin B. The AUC of the ROC curve showed better performance for 4.0 μg/mL, with values of 0.9671 and 0.9568 for colispot and polyspot, respectively. The Kappa index was 0.9305 for colispot and 0.9079 for polyspot. All isolates analyzed by VITEK ® 2 exhibited ROC curves with AUC values of 0.9541 and a Kappa index of 0.9049. Colispot and polyspot at a concentration of 4.0 μg/mL had higher results than the VITEK ® 2 system. The results showed that colispot and polyspot can be used with confidence in the practice of clinical laboratories.
Keywords: Polyspot; Colispot; Polymyxins; Resistance; Broth micro dilution; Detection method
Introduction
Infections due to Multidrug-Resistant Gram-Negative (MDR-GN) bacteria, especially carbapenem-resistant isolates, are increasingly reported in health care facilities and are responsible for nosocomial infections that may lead to fatal outcomes due to limited therapeutic options [1-3]. Carbapenem-resistant gram-negative bacterial infections remain a significant challenge associated with morbidity and mortality worldwide [4,5]. Acinetobacter baumannii and Pseudomonas aeruginosa, as well as Klebsiella pneumoniaecarbapenemase (KPC)- producing K. pneumoniae, are among the most common MDR-GN microorganisms now encountered in the clinical setting, and treating and managing them is a challenging task [6].
The increasing prevalence of MDR-GN has led to multiple reports regarding cases that required the clinical use of polymyxins [7]. These typically involve critically ill patients where the polymyxins are used as salvage therapy, either alone or in combination. The increasing use of colistin in human medicine and the recent discovery of plasmid- mediated polymyxin resistance highlight the need for reliable methods for polymyxin susceptibility testing [8].
The Clinical Laboratory Standard Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) recently gathered in a joint subcommittee and chose the Broth Micro Dilution (BMD) method as the reference method [9]. However, manufacturing BMD panels is laborious in routine clinical laboratories, and the commercial BMD panels available are expensive for many hospitals [10]. Automated dilution methods such as those performed by the BD Phoenix system could be an alternative for screening colistin resistance in laboratories that cannot perform manual BMD [11].
Recently, the Brazilian Ministry of Health published a National Action Plan for Prevention and Control of Antimicrobial Resistance in the healthcare field (PAN-BR). One of the guidelines is the improvement of surveillance and monitoring of MDR-GN in the field of human health to guide clinical treatment protocols and assess epidemiological trends. Therefore, due to the worldwide spread of resistance to polymyxins, including antimicrobials that act through chromosomal and plasmid mechanisms, phenotypic screening methods are required for routine laboratory detection [12].
In the clinical laboratory, the bacterial resistance of the antimicrobial class of polymyxins is underestimated, probably due to the challenges of the available easy methods to detect resistance to polymyxins. In addition, susceptibility testing guidelines do not have established cut-off points for the diffusion (Kirby-Bauer) technique for the detection of polymyxin resistance. Small clinical laboratories often do not have automated systems to analyze antimicrobial susceptibility, and BMD is laborious. In this context, we identified the importance of a screening test to identify resistance to polymyxins.
In this study, we evaluated the performance of the screening tests colispot [13] and polyspot (polymyxins B), described in this study. These tests can be deployed to detect resistance to polymyxins in laboratories during routine antimicrobial susceptibility testing by the agar dilution test (Kirby-Bauer). We also compared the performance of these news tests with the VITEK ® 2 automated system to determine colistin susceptibility and with the reference method BMD.
Methodology
Bacterial isolates
This study was carried out using consecutive clinical isolates of A. baumannii, P.aeruginosa and Enterobacteriaceae isolated between April and December 2018. In addition, 57 polymyxin resistant K. pneumoniae, previously characterized during 2016-2017, were included in the collection of tested strains. The isolates were identified using VITEK ® 2, and susceptibility profiles were determined with an AST 239/238 card. The interpretation was performed according to the CLSI 2018 criteria. The samples were stored in TSB (tryptic soy broth) with 15% glycerol (-80°C) until the tests were carried out.
Reference antimicrobial susceptibility testing
The BMD method was performed according to the EUCAST/CLSI joint guidelines. Briefly, BMD panels were prepared extemporaneously in 96-well sterile polystyrene microplates (Cralplast, Brazil). Dilutions of colistin (Sigma, colistin sulfate Salt) and polymyxin B (Bedfordpoly-B, polymyxin B sulfate) ranging from 0.125 to 128 mg/L were made in cation-adjusted MH broth (BBLTM, France), without the addition of polysorbate 80 (Tween 80) and with a final concentration of 5 × 105 CFU/mL bacteria in each well. This procedure was performed during microbiological routine, and the Minimum Inhibitory Concentrations (MICs) were read after 16 to 20 hours of incubation at 35 ± 2°C in ambient air. Escherichia coli ATCC 25922 and P. aeruginosa ATCC 27853 were included as control strains [14].
Colispot and polyspot
The purpose of the tests was to introduce a screening test that could
identify polymyxin resistance during routine microbiological testing. The tests were performed as described with some modifications [13]. Colistin and polymyxin B solutions were prepared at concentrations of 2.0, 4.0 and 8.0 mg/L in cation-adjusted MH broth according to the EUCAST/CLSI guidelines and stored at -20°C until use. The bacterial inoculum used was the same as that prepared during the VITEK ® 2 routine, with a final concentration of 5 × 105 CFU/mL bacteria. The bacterial suspension was spread on a petri dish containing Mueller- Hinton Agar (MHA) medium. Then, 10 μL of colistin and polymyxin B were dispensed on the surface of the medium at concentrations of 2.0 µg/mL, 4.0 μg/mL and 8.0 µg/mL and incubated at 35°C ± 2°C for 18 to 24 h. The MIC was defined as the lowest concentration of antibiotic capable of inhibiting the growth of the microorganism. Escherichia coli ATCC25922 and P. aeruginosa ATCC 27853 were included as control strains.
Statistical analysis
Statistical analysis was performed using the computing environment R (R Development Core Team 2018). We used the pROC and epiR packages to plot the Receiver Operating Characteristic (ROC) curves and to calculate the area under the ROC curve (AUC), and the concordance analysis was performed by Cohen's kappa coefficient.
Discrimination was evaluated from the AUC by comparing the two AUCs by DeLong's test. The non-parametric bootstrap method can be used to simulate real-world situations as well as to validate the tests. From the data obtained from the present study, we randomly generated 10000 bootstrap samples of the same size, obtained by resampling and replacing each sample, to validate the test. Tests were 2-tailed, and an alpha level of 0.05 indicated statistically significant results.
Breakpoints and definitions
The CLSI interpretive guidelines used for each antimicrobial tested were (μgmL) as follows: for P. aeruginosa, ≤ 2.0 μg/mL susceptible, 4.0 µg/mL intermediate, ≥ 8.0 μg/mL resistant (polymyxin B) and ≤ 2.0 μg/ mL susceptible, ≥ 4.0 µg/mL resistant (colistin) and for Acinetobacter spp., ≤ 2.0 μg/mL susceptible, ≥ 4.0 µg/mL resistant (for polymyxin B and colistin). Enterobacteriaceae was evaluated in the presence of colistin and polymyxin B using the EUCAST 2018 guidelines, with breakpoints and the following interpretation: ≤ 2.0 µg/mL susceptible, >2.0 μg/mL resistant.
Very major errors were defined when a value was determined as false susceptible, with an acceptable performance rate of 3%. Major errors were defined as a false-resistant result, with an acceptable performance rate of 3% [15].
If no bacterial growth was observed in any of the positions that were inoculated with the colispot and polyspottests at concentrations of 2.0 μg/mL, 4.0 μg/mL and 8.0 μg/mL, the result of colistin and polymyxin B was likely ≤ 2.0 μg/mL. If the bacteria grew only in the position with the colistin and/or polymyxin B test at a concentration of 2.0 μg/mL, the probable result of colistin and/or polymyxin B was 4.0 μg/mL. If bacterial growth occurred at the positions with colistin and/or polymyxin B at concentrations of 2.0 μg/mL and 4.0 μg/mL, the probable result was 8.0 μg/mL. If bacterial growth was observed in the presence of colistin and/or polymyxin B at concentrations of 2.0 μg/mL, 4.0 μg/mL, and 8.0 μg/mL, the likely result of colistin and/or polymyxin B was ≥ 16.0 μg/mL.
Results
We analyzed 521 microorganisms isolated during a series of clinical studies and a collection of 57 polymyxin-resistant K. pneumoniae. A total of 578 bacterial strains were used in this study, including A. baumanii 244 (42.2%), P. aeruginosa 108 (18.7%), K. pneumoniae 214 (37%), E. coli 3 (0.5%) and Enterobacter spp. 9 (1.6%).
In the evaluation of the BMD method for all microorganisms studied, 420 isolates were colistin sensitive and 158 were colistin resistant. The BMD method for K. pneumoniae detected 68 isolates that were susceptible to colistin and 146 isolates that were resistant to colistin. For polymyxin B, the BMD method identified 423 polymyxin B-sensitive isolates and 155 polymyxin B-resistant isolates. Furthermore, for K. pneumoniae, 70 isolates were susceptible to polymyxin B and 144 were resistant to polymyxin B. In addition, the performance of the VITEK ® 2 automated system identified 416 isolates that were susceptible to colistin and 145 isolates that were resistant to colistin. For only K. pneumoniae, 65 of the isolates were susceptible to colistin and 139 were resistant to colistin, as shown in Table 1.
| Drug | Microorganism | Method | No. of susceptible | No. of resistant | ME (%) | VME (%) |
|---|---|---|---|---|---|---|
| Colistin | All cases (n=578) | BMD | 420 | 158 | NA | NA |
| VITEK ® 2 | 429 | 149 | 0.69 | 2.24 | ||
| Colispot test | ||||||
| 2.0 µg/mL | 376 | 202 | 8.3 | 0.69 | ||
| 4.0 µg/mL | 418 | 160 | 4.34 | 1.21 | ||
| 8.0 µg/mL | 440 | 138 | 4.32 | 0.86 | ||
| k.pneumoniae (n=214) | BMD | 68 | 146 | NA | NA | |
| VITEK ® 2 | 65 | 139 | 2.33 | 2.33 | ||
| Colispot test | ||||||
| 2.0 µg/mL | 59 | 155 | 5.14 | 0.93 | ||
| 4.0 µg/mL | 69 | 145 | 1.86 | 2.33 | ||
| 8.0 µg/mL | 86 | 127 | 1.07 | 9.81 | ||
| Polymyxin B | All cases (n=578) | BMD | 423 | 155 | NA | NA |
| Polyspot test | ||||||
| 2.0 µg/mL | 389 | 189 | 8.98 | 0.69 | ||
| 4.0 µg/mL | 420 | 158 | 2.07 | 1.55 | ||
| 8.0 µg/mL | 438 | 140 | 1.03 | 3.63 | ||
| k.pneumoniae (n=214) | BMD | 70 | 144 | NA | NA | |
| Polyspot test | ||||||
| 2.0 µg/mL | 61 | 153 | 5.6 | 1.4 | ||
| 4.0 µg/mL | 70 | 144 | 2.8 | 2.8 | ||
| 8.0 µg/mL | 85 | 129 | 1.4 | 8.41 | ||
Table 1: Performance characteristics of colistin and polymyxin B susceptibility testing methods in comparison to broth microdilution of 578 isolates BGN.
The isolates were designated as resistant based on broth micro dilution. A MIC >2 µg/mL was considered resistant.
The results of the activity of colispot and polyspot at concentrations of 2.0 μg/mL, 4.0 μg/mL and 8.0 μg/mL and the Very Major Errors (VME) and Major Errors (ME) are shown in Table 1. The K. pneumoniae isolates were separated for analysis because they presented a higher frequency of resistant isolates in our laboratory.
The test performances of colispot and polyspot were measured by the AUC of the ROC curve, sensibility, specificity, Positive Predictive Value (PPV), Negative Predictive Value (NPV) and the Kappa index, as shown in Table 2.
| Drug | Method | AUC (CI 95%) |
Sensibility (CI 95%) | Specificity (CI 95%) |
PPV (CI 95%) |
NPV (CI 95%) |
Correctly classified cases (CI 95%) |
Kappa (SE) |
|---|---|---|---|---|---|---|---|---|
| Colistin | Colispot test | |||||||
| 2.0 µg/mL | 0.9302 (0.9106-0.9498) |
0.9893 (0.9798-0.9970) |
0.7623 (0.6975-0.8192) |
0.883 (0.8483-0.9122) |
0.9559 (0.9113-0.9821) |
0.91 (0.8836-0.9320) |
0.7916 (0.0419) |
|
| 4.0 µg/mL | 0.9671 (0.9496-0.9847) |
0.9832 (0.9580-0.9924) |
0.9437 (0.8959-0.9735) |
0.9785 (0.9597-0.9901) |
0.9556 (0.9108-0.9820) |
0.9723 (0.9554-0.9840) |
0.9305 (0.0415) |
|
| 8.0 µg/mL | 0.9149 (0.8859-0.9439) |
0.9431 (0.9172-0.9628) |
0.9637 (0.91740.9881) |
0.988 (0.9724-0.9961) |
0.8417 (0.7753-0.8949) |
0.948 (0.9267-0.9647) |
0.8639 (0.0414) |
|
| VITEK ® 2 | 0.9541 (0.9321-0.9761) |
0.9669 (0.9487-0.9837) |
0.9731 (0.9326-0.9926) |
0.9904 (0.9757-0.9973) |
0.9177 (0.8634-0.9554) |
0.9705 (0.9533-0.9827) |
0.9246 (0.0415) |
|
| Polymyxin B | Polyspot test | |||||||
| 2.0 µg/mL | 0.9422 (0.9237-0.9607) |
0.9897 (0.9738-0.9971) |
0.7989 (0.7346-0.8536) |
0.9101 (0.8787-0.9920) |
0.9741 (0.9352-0.9816) |
0.9273 (0.9030-0.9471) |
0.8268 (0.0412) |
|
| 4.0 µg/mL | 0.9568 (0.9367-0.9769) |
0.9785 (0.9597-0.9900) |
0.924 (0.8710-0.9601) |
0.9716 (0.9509-0.9852) |
0.9225 (0.8686-0.9593) |
0.9636 (0.9449-0.9737) |
0.9079 (0.0415) |
|
| 8.0 µg/mL | 0.9142 (0.8853-0.9432) |
0.952 (0.927-0.9700) |
0.9532 (0.9090-0.9841) |
0.9858 (0.9638-0.9947) |
0.8645 (0.8038-0.9141) |
0.9532 (0.9307-0.9675) |
0.8772 (0.0414) |
|
Table 2: Performance characteristics of colistin and polymyxin B susceptibility testing methods in comparison to broth microdilution, of 578 isolates BGN.
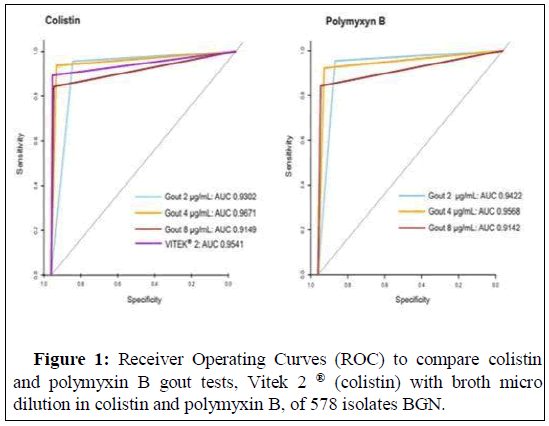
As shown in Figure 1, Receiver Operating Characteristics (ROC) curves of colispot, polyspotand VITEK ® 2 (colistin) were determined by the broth microdilution method in the presence of colistin and polymyxin B, for all isolates studied, which showed that the AUC had the best predictive ability for the 4.0 μg/mL colispot and polyspot tests according to the DeLong test (Table 3).
| Drug | Methodology | G out 2.0 µg/mL | G out 4.0 µg/mL | G out 8.0 µg/Ml | VITEK ® 2 |
|---|---|---|---|---|---|
| Colistin | Colispot test | ||||
| 2.0 µg/mL | - | p<0.0001 | p=0.3228 | p=0.065 | |
| 4.0 µg/mL | - | - | p<0.0001 | p=0.285 | |
| 8,0 µg/mL | - | - | - | p=0.0192 | |
| VITEK ® 2 | - | - | - | - | |
| Polymyxin B | Polyspot test | ||||
| 2.0 µg/mL | - | p=0.1129 | p=0.1211 | - | |
| 4.0 µg/mL | - | - | p=0.0203 | - | |
| 8,0 µg/mL | - | - | - | - | |
Table 3: DeLong test results of Colispot and Polyspot test and VITEK ® 2 area under the ROC curve (AUC), of 578 isolates BGN.
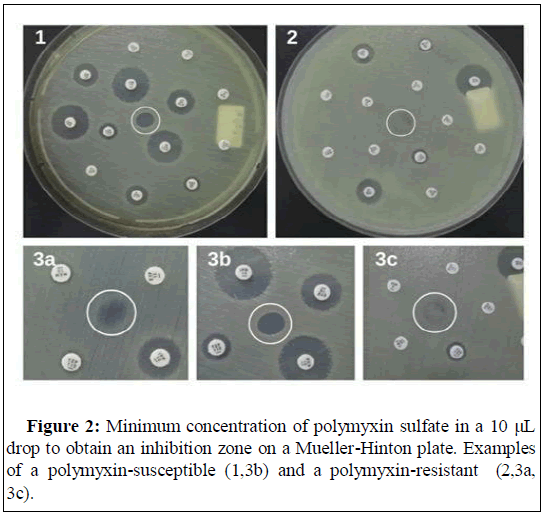
In Figure 2, show the application of colispot and polyspot to evaluate polymyxin resistance antimicrobial susceptibility test during microbiological routine.
Discussion
Since the emergence of intrinsic and transferable mechanisms of resistance to polymyxin is becoming a critical issue throughout the world, the development of rapid and reliable methods for determining susceptibility and resistance to polymyxins is an urgent need of clinical laboratories [16,17]. Thus, the European Center for Disease Prevention and Control (ECDC) published a document in June 2016 calling for a rapid risk assessment to control the spread of plasmidmediated colistin resistance in Enterobacteriaceae microorganisms [18]. Thus, the PAN-BR also presented a strategic plan that aims to ensure that the capacity to treat and prevent infectious diseases with safe and effective drugs is maintained, which requires the promotion of research to gain knowledge of antimicrobial resistance [12]. CLSI and EUCAST have referenced the BMD method as a reference method for susceptibility testing. This gold standard method is difficult to perform in routine laboratories because it requires skilled researchers, is time consuming, and requires the manual preparation of antibiotic solutions, making the detection of clinical isolates with resistance to polymyxins in laboratories difficult [19].
This study proves through statistical tests by the agar dilution method (colispot and polyspot) can be used to screen and predict resistance to polymyxins. In the analysis of the isolates together, the sensitivity and specificity tests indicate high precision. We plotted a Receiver Operating Characteristic (ROC) curve to examine the true positive rate (sensitivity) versus false positive rate (specificity) as a measure of the inherent validity of our screening test for colistin and polymyxin B. Using the ROC analysis, it was evident that for the colispot and polyspot tests, the AUC exhibited a higher predictive capacity at a concentration of 4.0 μg/mL than at concentrations of 2.0 or 8.0 μg/mL, with comparable accuracy based on the DeLong test. Colispot and polyspot values at concentrations of 4.0 μg/mL showed the best results for VME and ME.
To validate the new screening test, we also analyzed the Kappa concordance index, which presented values higher than the recommended definitions at all test concentrations. However, colistin and polymyxin B at a concentration of 4.0 μg/mL demonstrated a better agreement, which proved to be a methodology with good performance and is therefore reliable in clinical laboratories.
In our study, the VITEK ® 2 systems also presented relevant statistical results when analyzed for all 578 Gram negative bacillus (BGN) isolates. The AUC value indicated high sensitivity and specificity of the system and VME values below the acceptable limit. The Kappa concordance coefficient presented good accuracy, affirming that it is a safe methodology for screening and detecting colistin susceptibility. However, previous studies have shown that the colistin test of Enterobacteriaceae isolates performed by the VITEK ® 2 system had a high VME rate (36%) [20]. In addition, in a study of 361 BGN isolates, 2.2% (ME) and 6.3% of VME were observed. The VITEK ® 2 automated system showed variable performance among BGN species for detecting colistin susceptibility [10].
The limitations of our study occurred with some microorganisms in which resistance to polymyxins were not frequent. Thus, the VME and ME results remained outside the desired limits for good test performance, most likely due to the low degree of freedom of these isolates.
Although all tests that are recommended to validate a methodology showed excellent results, such as the AUC and the Kappa concordance index. The DeLong test was performed in our study to strengthen the validation of the data. The test indicated statistically significant results for the gout test, mainly for 4.0 μg/mL gout.
Based on a previous study on colispot test inspired by a technique used and polyspot, in this study, the authors conclude that these tests are particularly robust since they can be used with the main standards used for agar dilution tests (Kirby-Bauer) [13,21].
Conclusion
In this study, we observed that the concentration of 4.0 μg/mL, for colispot and polyspot, had similarly higher results than the VITEK ® 2 system and can be used with confidence in the practice of clinical laboratories. We emphasize that the colispot and polyspot tests are an alternative and simple phenotypic method for screening and is an easy- to-interpret and cost-effective technique that can be easily implemented in clinical microbiological practice to detect resistance to polymyxins. In summary, we emphasize the need for the implementation of easy and inexpensive tests in routine clinical microbiology laboratories to detect and contain the spread of polymyxin-resistant microorganisms.
References
- Vincent JL, Rello J, Marshall J, Silva E, Anzueto A, et al. (2009) International study of the prevalence and outcomes of infection in intensive care units. Jama 302: 2323-2329.
- Wright H, Bonomo RA, Paterson DL (2017) New agents for the treatment of infections with gram-negative bacteria: restoring the miracle or false dawn? Clin Microbiol Infect 23: 704-712.
- Shin B, Park W (2017) Antibiotic resistance of pathogenic acinetobacter species and emerging combination therapy. J Microbiol 55: 837-849.
- Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, et al. (2013) Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13: 785-796.
- Van Duin D, Perez F, Rudin SD, Cober E, Hanrahan J, et al. (2014) Surveillance of carbapenem-resistant Klebsiella pneumoniae: Tracking molecular epidemiology and outcomes through a regional network. Antimicrob Agents Chemother 58: 4035-4041.
- Scheld M, Talbot GH, Gilbert D, Edwards JE, Bradley JS, et al. (2008) Bad bugs, no drugs: No eskape! An update from the infectious diseases society of America. Clin Infect Dis 48: 1-12.
- Karlowsky JA, Draghi DC, Jones ME, Thornsberry C, Friedland IR, et al. (2003) Surveillance for antimicrobial susceptibility among clinical isolates of pseudomonas aeruginosa and acinetobacter baumannii from hospitalized patients in the United States, 1998 to 2001. Antimicrob Agents Chemother 47: 1681-1688.
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, et al. (2016) Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect Dis 16: 161-168.
- European Centre for Disease Prevention and Control (ECDC) (2016) Rapid risk assessment. Plasmid-mediated colistin resistance in enterobacteriaceae. 1-14.
- Girardello R, Cury AP, Franco MRG, Di Gioia TR, de Almeida JN, et al. (2018) Colistin susceptibility testing and Vitek-2TM: is it really useless? Diagn Microbiol Infect Dis 91: 309-311.
- Jayol A, Nordmann P, Lehours P, Poirel L, Dubois V (2018) Comparison of methods for detection of plasmid-mediated and chromosomally encoded colistin resistance in enterobacteriaceae. Clin Microbiol Infect 24: 175-179.
- Brazil (2018) Ministry of health national action plan for the prevention and control of antimicrobial resistance in the scope of Unified Health 2018-2022. Brasilia, Brazil: Ministry of Health 10.
- Jouy E, Haenni M, Le Devendec L, Le Roux A, Chatre P, et al. (2017) Improvement in routine detection of colistin resistance in E. coli isolated in veterinary diagnostic laboratories. J Microbiol Methods 132: 125-127.
- CLSI (2018) Performance standards for antimicrobial susceptibility testing. CLSI supplement M100. Wayne PA. Perform stand antimicrob susceptibility testing CLSI Suppl M100 Wayne PA: 282.
- Simar S, Sibley D, Ashcraft D, Pankey G (2017) Colistin and Polymyxin B minimal inhibitory concentrations determined by Etest found unreliable for gram-negative bacilli. Ochsner J 17: 239-242.
- Caspar Y, Maillet M, Pavese P, Francony G, Brion JP, et al. (2017) Mcr-1 colistin resistance in esbl-producing Klebsiella pneumoniae, France. Emerg Infect Dis 23: 874-876.
- Dona V, Bernasconi OJ, Kasraian S, Tinguely R, Endimiani A (2017) A SYBR ® green-based real-time PCR method for improved detection of mcr-1-mediated colistin resistance in human stool samples. J Glob Antimicrob Resist 9: 57-60.
- European Centre for Disease Prevention and Control (ECDC) (2016) Rapid risk assessment. Plasmid-mediated colistin resistance in enterobacteriaceae improved laboratory methods for colistin resistance.
- Le Marechal M, Fressard L, Agrinier N, Verger P, Pulcini C (2018) General practitioners perceptions of vaccination controversies: French nationwide cross-sectional study. Clin Microbiol Infect 24: 858-864.
- Chew KL, La M-V, Lin RTP, Teo JWP (2017) Colistin and polymyxin B susceptibility testing for carbapenem-resistant and mcr-positive enterobacteriaceae: Comparison of sensititre, microscan, vitek 2, and etest with broth microdilution. J Clin Microbiol 55: 2573-2582.
- Halling SM, Jensen AE, Olsen SC (2008) Defensin susceptibility and colonization in the mouse model of AJ100, a polymyxin B-resistant, brucella abortus RB51 isolate. Curr Microbiol 56: 274-278.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 2168
- [From(publication date): 0-2021 - Jul 04, 2025]
- Breakdown by view type
- HTML page views: 1559
- PDF downloads: 609