Research Article Open Access
Performance of Isolated Kocuria sp. SAR1 in Light Crude Oil Biodegradation
| Abdullah M. El Mahdi1, Hamidi Abdul Aziz1,2*, Salem S Abu Am1, Nour Sh. El-Gendy3 and Hussein Nassar3 | |
| 1School of Civil Engineering, Engineering Campus, Universiti Sains Malaysia, 14300 NibongTebal, Penang, Malaysia | |
| 2Solid Waste Management Cluster, Engineering Campus, Universiti Sains Malaysia, 14300 Penang, Malaysia | |
| 3Egyptian Petroleum Research Institute, Nasr City, Cairo 11727, Egypt | |
| Corresponding Author : | Prof. Hamidi Abdul Aziz School of Civil Engineering Engineering Campus Universiti Sains Malaysia 14300 Nibong Tebal, Penang Malaysia Tel: + 60-45996215 Fax: +60-45941009 E-mail: cehamidi@eng.usm.my |
| Received June 09, 2015; Accepted July 06, 2015; Published July 08, 2015 | |
| Citation: El Mahdi AM, Aziz HA, Abu Am SS, El-Gendy NS, Nassar H (2015) Performance of Isolated Kocuria sp. SAR1 in Light Crude Oil Biodegradation. J Bioremed Biodeg 6:303. doi:10.4172/2155-6199.1000303 | |
| Copyright: © 2015 El Mahdi AM, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
In the current study; Kocuria sp. SAR1 was isolated from ‘Tobruk Refinery’ oil water pit, located along the eastern coast of Libya. The isolated bacterial strain SAR1 was characterized as an aerobic, Gram +ve, spherical-shaped, oxidase – but catalase +. Phenotypic characters and phylogenetic analysis based on the 16S rRNA gene of the isolate SAR1 showed that it was related to members of the Kocuria genus. The alignment of the 16S rRNA gene sequences of SAR1 with sequences obtained by doing a Blast searching revealed 96% similarity to Kocuria palustris strain TAGA27. Solid waste dates (SWD) and corn steep liquor (CSL) as agro-industrial products were performed to enhance the performance of Kocuria sp. SAR1 in crude oil biodegradation. During bacterial growth, high emulsifying activity to the presence of cells was observed, which is concluding the production of bio surfactant by strain SAR1. The bacterial strain showed removal ef?ciency of 68% and 70% of crude oil in 28 days when cultivated with 0.2% (w/v) of CSL and SWD, respectively. Crude oil metabolizing bacterium can secrete surfactants using agro industrial as substrates, which further enhance the hydrocarbon degradation.
| Keywords |
| Isolation; Identification; 16S Rdna; Biodegradation; Crude oil; SWD |
| Introduction |
| The ocean and other water bodies have captured the imagination of people for thousands of years. Most of the life forms exist mostly in these water bodies and therefore, specific laws, and regulations have been framed to protect this delicate marine environment. It is also the major source of food chain and is known for diversity of aquatic species [1,2]. Marine oil pollution occurs when any organic or toxic chemical substances enter the sea water. These chemical hazards can lead to severe pollution of the system either for short duration or over long period of time. Primarily it affects the biological process thereby damaging the marine life cycle. Most common causes of marine pollution are due to oil spill occurring at sea. These oil spills deteriorate the marine environment to such an extent that, it destroys the existing ecosystem accordingly affecting the bio diversity and human wellbeing [3-5]. The oil spill results in significant changes in its physical and chemical structure [6,7]. The photolysis of oil can result in the formation of many byproducts such as the aromatic oxygenated compounds, aliphatic, benzoic and naphthanoic acids, alcohols, phenols and aliphatic ketones [8,9]. |
| In Libya, 5 oil terminal facilities and many different operating companies are discharging effluents at risk rate. Oil pollution presents the hazard along parts of the Libyan Marine Coast where Oil Industries are located. This severely affects ecosystem. With the focus on the protection of environment and pollution control in Libya, most of the terminal facilities with the conventional ballast water process are likely to face a serious threat in coming years and need has been felt for a safe, environmental friendly process which eliminates the use of discharging of ballast water into sea. At present, bioremediation use of microorganisms to remove pollutants is often the most suitable method for remediation of especially petroleum hydrocarbons, because it is cost-effective and, it converts the petroleum hydrocarbons into the harmless by-products such as carbon dioxide and water [10,11]. Various microbial populations, including Bacteria, Fungi and Algae can metabolize the hydrocarbons found in crude oil. In literature, bacterium was considered to be the most important group of petroleum degrading organisms because its allow adaptation to various environments, have a rich taxonomic, metabolic, physiological, and it has more rapid metabolic rates. Moreover, bacteria can be genetically manipulated to improve their bioremediation capabilities [12,13]. There are at least 175 genera of bacteria that can metabolize petroleum hydrocarbons, which include- Pseudomonas, Aeromonas, Bacillus, Flavobacterium, Corynebacterium, Micrococcus etc. [14]. Based on crude oil degradation capacity, Pseudomonas aeruginosa is the most active hydrocarbon utilize in crude oil. Previous observations have identified the Pseudomonas genus most efficient among hydrocarbon degrading microorganisms [15-17]. |
| ¨Further use of surfactants has been found to enhance degradation of crude oil [18,19]. Among various surfactants, rhamnolipids are considered to be the most efficient way in degrading hydrocarbons [20-22]. Most studies reported the effectiveness of using consortium bacterium compared with the single strain bacteria. Single strain is still not effective in terms of biodegradability. Both consortium and single strains reported limited performance when used in high crude oil concentration. The isolated Kocuria sp. from contaminated water by crude oil was recently received attention in crude oil biodegradation [23]. However, the success of their production depends on the increase of yield, the development of economic biotechnology processes, and the use of low cost effective renewable agro-industrial substrates for their production. However, the performance of employing SWD and CSL as a low-cost material to enhance single strain bacteria in removing highly contaminated crude oil was not well investigated. The aim of this study is to evaluate the effectiveness of isolated Kocuria sp. on crude oil biodegradation. |
| Materials and Methods |
| Sampling |
| In the present study, Oil contaminated water samples were collected from Tobruk Refinery (Libya) Figure 1, where Arabian Gulf Oil Co. Company (AGOCO.) It has produced huge amount of crude oil. Samples were collected in an autoclaved vial by using simple sampling technique. Care was taken in handling and sampling to avoid contamination of the samples and returned to the laboratory for bacterial isolation immediately. If necessary, the sample vials were held under refrigeration at 4°C until isolation, which was no later than 48 hours after sampling. |
| Isolation of hydrocarbon-degrading bacteria |
| Collected samples were serially diluted and plated onto a basal medium amended with 0.5% (w/v) crude oil hydrocarbon as the sole carbon source. Flasks were incubated at 30°C on a rotary shaker (150 rpm). 10 mL from each flask was transferred separately to fresh flasks containing 90 mL of Enrichment media, and the procedure was repeated for three transfers namely; 7, 14, and 28 days, respectively. Serial dilutions (10-7) of each transfer were inoculated on TGY medium plates to enumerate TCFU and onto BSM-Crude Oil plates to count BDM. Plates were incubated at 30°C, and colonies were enumerated after 48 h on TGY plates and after 168 h on BSM-Crude Oil plates. Separate colonies from BSM-Crude Oil plates were picked and purified on BSM-Crude Oil plates. Individual colonies were streaked successively onto the same medium (basal medium with 0.5 (w/v) crude oil hydrocarbons for isolation and purification [24]. The basal Salt Mineral (BSM) medium according to Piddington CS [25], was prepared by dissolving 2.44 g KH2PO4, 5.57 g NaHPO4. 2.0 g NH4Cl, 0.2 g MgCl2.6H2O, 0.001 g FeCl3.6H2O, 0.001 g CaCl2.2H2O in one liter sea water and pH was adjusted to 7.0 using 10% NaOH, blended with 0.5% (w/v) Sarir crude oil was used as carbon source and autoclaved for 20 minutes at 120°C. For isolation and enumeration of total viable cells; BSM with crude oil 0.5% (w/v) and TGY were solidified by 2% agar and plates were used. Each medium was autoclaved for 20 minutes at 120°C. According to Benson HJ [26] Trypyon glucose yeast extracted medium (TGY) was prepared by dissolving 5.0 g Trypton, 3 g yeast extracts, and 1 g glucose in one liter double distilled water and autoclaved. For the total viable count; each medium pH was adjusted to 7.0 using 10% NaOH. |
| Local Solid waste date (SWD) was purchased from Benghazi local market, Libya, and its compositions were (as reported by manufacturer, for each 100 ml : Moisture, 13-16%; Protein, 1.0-2.3%; Ash, 1.4-1.8%; total sugar, 75-76% (Glucose: 38%; Fructose: 36%) - Pectin, 0.1- 0.2%; Tannin, 0.2-0.3%; pH, 4.5-5.3; TDS, 74-75%. Corn steep liquor (CSL) used for this study was provided from El-Nasr Pharmaceutical Company; carbon: nitrogen: phosphorus content of CSL was 6.8: 1.06: 2.14 (% w w−1), respectively. |
| Identification and characterization of bacteria |
| Biochemical characterization: To identify and characterize the bacteria isolates, the designed Strain SAR1 colonies were identified by a combination of information from primary and secondary identification. Morphological, physiological and biochemical characteristics of pure isolated were examined according to the Bergey’s Manual of Determinative Bacteriology. Primary identification was done based on colony and cell morphology and Gram staining [27]. Representative colonies of strain SAR1 appeared on plates were checked for purity through the microscopy and pure isolates were streaked on slants of BSM-Crude Oil medium on which they developed during isolation and stored at -40°C for further investigation. For secondary identification, Bacterial isolates was characterized by 16S rRNA gene sequencing [28]. After identification the isolate was deposited in the culture collection for long-term preservation. DNA isolation and amplification were done in Sigma, as follows; |
| a. DNA Isolation: The most efficient biodegrading bacterial isolate was incubated separately in TGY for 48 h at 30°C on a rotatory shaker (150 rpm). The genomic DNA was extracted using Gene JET Genomic DNA Purification Kit K0729 (Fermentas, USA). |
| b. PCR Amplification of 16S rDNA Regions: Two sets of primers were used to amplify regions specific for almost all eubacteria 16S rDNA sequences. The 16SrDNA gene was amplified by polymerase chain reaction (PCR) using the following primers [28]. The universal primers Fd1 and Rd1 (Fd1,5-TGCCTGGTAGTGGGGGATAA and Rd1,3’- CCAGGTAAGGTTCTCGCGTT-3’). |
| The reaction was prepared with 0.5 μL of Dream-Taq 5 U/μL (fermentas, USA), 5 μL of Dream-Taq Buffer 10x, 5 μL of target DNA, 1 μL of dNTP each 20 mM, 1 μL of each appropriate primer 10 pmol/ μL and 36.5 μL dH2O were added. The final reaction volume was 50 μL. |
| The PCR condition was as follows: |
| Step 1: Pre-Denaturation: 94°C 5 min. |
| Step 2: 35 cycles of: |
| - Denaturation 94°C 40 sec. |
| - Annealing 48°C 40 sec. |
| - Extension 72°C 3 min. |
| - Final Extension 72°C 10 min |
| Step 3: Hold at: 4°C Indefinitely |
| After completion of PCR program, the visualization of PCR products was carried out using, 5 μL of the suspension electrophoresed on 1% agarose gels in 1X Tris-Acetate EDTA (TAE) buffer, which were then stained with ethidium bromide and examined under UV light. Bands were excised, and DNA was purified from gel slices using QIAquick Gel Extraction Kit, Cat. No. 28704 (Qiagen, USA). The purified PCR products were sequenced with the same primer that has been used in amplification of the target sequence. Sequencing was done by an ABI 3730 XL automatic DNA sequencer at Sigma, Giza, Egypt. The 16S rDNA sequences (Query sequence) were initially analyzed at NCBI server (http://www.ncbi.nlm.nih.org) using BLAST tool (http:// blast.ncbi.nlm.nih.gov/Blast.cgi) and corresponding sequences from database were downloaded. Evolutionary history was inferred using the Neighbor-joining method [29]. The tree was drawn to the scale, with branch lengths in the same units as those of the evolutionary distance used to infer the phylogenetic tree. |
| Crude oil-biodegradability assay: Biodegradation assays were also performed in liquid culture to examine the ability of isolated microorganisms to utilize crude oil as a sole energy source according to Ismail et al. [30]. This assay was done as follows: Cells were incubated at 30°C in TGY for 24 h in a shaking incubator (150 rpm). Next, cells were pelleted by centrifugation at 5000 rpm for 15 min and then washed three times with BSM. Washed cells were inoculated into BSM that contained crude oil as a sole energy source, to test the ability of isolates to degrade crude oil (Crude Oil-biodegradability assay). The inoculum was adjusted so that the beginning absorbance was (A600 0.3). The cultures were incubated at 30°C for seven days, in a shaking incubator (150 rpm). The growth was monitored at the prescribed time intervals by optical density at 600 nm, Using UV/Vis spectrophotometer (JASCO, V570, USA); non inoculated BSM was used as blank. The pH of the cultures was determined using pH-meter (DIGMED DM-22, Brazil). The residual of oil content in the culture flask was extracted by using a 1:1 proportion of organic solvent (n-hexane: BSM) followed by evaporating the solvent phase and measuring the weight of the dry extract. All the experiments were carried out in duplicate, and the mean values were considered. |
| Analytical study: Total petroleum hydrocarbons (TPH) were measured by the Environmental Protection Agency, (EPA Method-1664) [31] US-EPA [32], and serially extracted three times with analytical grade n-hexane (Merck, Darmstadt, Germany) in separator funnels. The extracts were dried over granular anhydrous sodium sulfate (Merck, Darmstadt, Germany). |
| The solvent was distilled from the extract and the n-hexane extractable material (HEM) was desiccated and weighed. The HEM was re-dissolved in n-hexane and silica gel was added to the solution containing the re-dissolved HEM to remove polar materials. |
| The solution was filtered to remove the silica gel, the solvent was distilled, and the silica gel treated n-hexane extractable material (SGT-HEM) is desiccated and weighed. Quality assurance and quality control were carried out according to the same procedure. Calibration verification and analysis of blanks were performed daily. Percent recovery (S) was calculated using Eq. (1): |
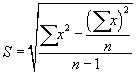 (1) (1) |
| where n is the number of samples and x is the percent of recovery in each sample. Moreover, matrix spike (MS) was tested to ensure the accuracy of the analysis. |
| The relative percent difference (RPD) between the matrix spike and matrix spike duplicated (MSD) were computed using Eq. (2): |
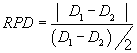 (2) (2) |
| where D1 is the concentration of hexane extractable material in the sample and D2 is the concentration of hexane extractable material in the duplicate sample. |
| Results and Discussion |
| Isolation and identification of the bacteria |
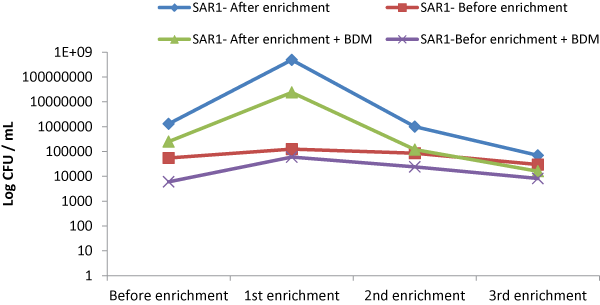
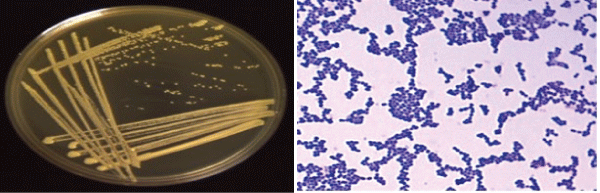
| Produced water sample were collected from the oil contaminated site showed density in the range of 1.3 × 109 CFU/ml to 1.7 × 1010 CFU/ ml. Subsequently, the final growth rate of 2.1 × 1010 CFU/ml to 1.1 × 1011 CFU/mL (CFU: colony forming unit) were measured for crude oil, enhanced with CSL and SWD respectively. Thus concluded the presence of visual emulsification was found to be growth associated in strain SAR1, where a parallel relationship exists between growth substrate utilization and emulsification activity. However, Xu et al. [33] reported that there was no decrease in the CFU of strain SY23 during the degradation even at higher concentration of crude oil. These results indicated that the bacterial strain SAR1 supported growth even at higher concentration. An enrichment culture initiated in basal medium containing 0.5% (w/v) crude oil as the carbon and energy source became turbid and the dark layer of crude oil became clear, indicating the degradation of this substrate. The obvious increase in the count of BDM and TCFU after Enrichment for one week in Enrichment medium, illustrated in Figure 1, might be due to the adaptation of the indigenous microbial populations. The viable counts of BDM either before or after Enrichment were less than TCFU indicating that; not all the indigenous microorganisms have the enzymatic system capable to degrade crude oil hydrocarbon and only microorganisms that have the required enzymatic system to metabolize crude oil would grow on the BSM/ containing crude oil. Enrichment culture showed the presence of aerobic bacteria with diverse cellular morphologies and then was used in the isolation of pure bacterial strains in solid media. One colony was picked, and a pure culture designed SAR1 was selected (Figure 2). |
| This culturing technique based upon the principle that when material-containing microorganism is cultured each viable microorganism will develop into a colony [34]. It is known that Gram (+) ve isolate designated as SAR3 belonged to the genus Kocuria. The Gram positive bacterium had spherical shape; opaque, moderate with entire edges occurred in groups. It was aerobic, oxidase -negative, but catalase- positive. Enrichment culture showed the presence of aerobic bacteria with diverse cellular morphologies, which was isolated in solid media to obtain pure bacterial strains as shown in Figure 2. |
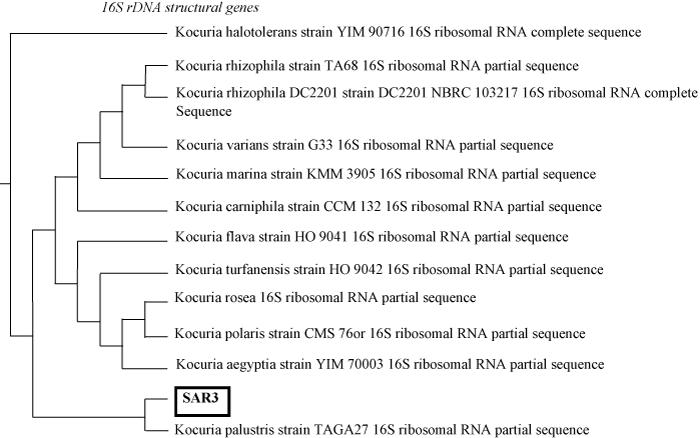
| The strain was identified as Kocuria sp. strain SAR1 and watersoluble pigments that gave it a characteristic yellow-orange. The selection of the strain was based on its high capacity to degrade crude oil, in solid and liquid media. The strain SAR1 was used for further characterization. This strain was first identified using classical biochemical morphological characteristics as shown in Figure 2. The molecular identification of isolates was performed amplifying and sequencing the 16S rRNA gene and comparing the sequences to the database of known 16S rRNA sequences. Nucleotide sequence of the 1160 bp fragments containing the Kocuria palustris strain TAGA27 1, 16S rDNA structural genes and deposited in the Genbank (accession number: NR-026451.1). The phylogenetic analysis suggested that the strain belonged to Kocuria palustris. and shows a higher 16S rRNA gene sequence similarity of 96% with Kocuria palustris strain TAGA271 summarized in Figure 3. |
| For the isolate, Gram staining, morphology analyses and the 16S rDNA sequence studies reveal, that this isolate belongs to Kocuria palustris strain TAGA27, based on literature available on crude oildegrading and emulsifying TAGA27 strain has not been reported before as isolate from oil-polluted sites in Libya. |
| Crude oil biodegradation |
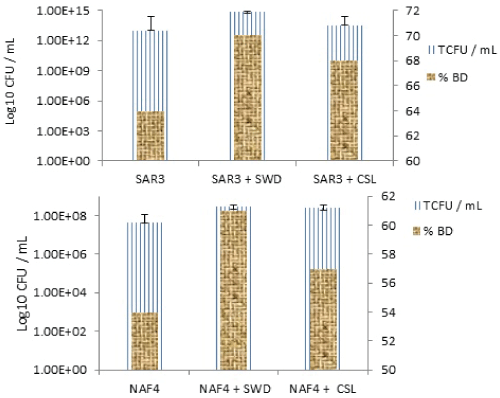
| The cultures were grown aerobically at 28°C for 28 days with constant shaking (120 rpm). During the experiment, the inoculated culture became turbid gradually due to mixing the crude oil with water and emulsification. Any changes in the physical condition through the medium were not observed during the control experiment, it remained clear, and dispersed crude oil on the water surface was as same as the beginning. Two of the isolated (NAF4, and SAR3) particularly grew well on the BSM, with minimum and maximum times showing dense growth by day 28, based on a preliminary study conducted (Table 1). However, the majority of the isolates in this study grew poorly or not on the BSM media at all despite many having been isolated on them. These isolated likely exhibited initial tolerance to these compounds but could not biodegrade or metabolize them efficiently enough to maintain their growth long term. This result indicates that indigenous microbial populations can biodegrade petroleum hydrocarbons in the polluted water sample, but the process is very slow. Table 1 demonstrates that oil removal by natural attenuation without nutrient supplementation or bacterial addition was much slower compared with combined bio augmentation and bio stimulation. On day 28, only about 38% of crude oil hydrocarbons was removed by natural attenuation. By contrast, during the same period for oil concentration, a value of 54% and 64% removal was obtained by single strains NAF4 and SAR3, respectively. 57%, 61% removal by the single strain NAF4 and 68%, 70% by single strain SAR3 by enhancing different organic nutrients as CSL and SWD 0.2% (w/v) concentration, respectively, which are not only acted as nutrients for the microbial growth but also act as an important source for isolation of potential bio surfactant producing microorganisms, respectively (Table 1). |
| Furthermore, the addition of SWD increased the degrading capability to 61% when added to single strain NAF4 and 70% when added to single strain SAR3. The addition of CSL improved single strain NAF4 and achieved 61% degradation, whereas single strain SAR3 could achieve 68% degradation. Single strain NAF4 achieved 54% degradation and increased to 57% and 61% when CSL and SWD were added, respectively. More than half of the oil in bio augmentation reactors was consumed by bacteria and nutrient concentration within 28 days, which was achieved because the well-adapted microbial cells were in direct contact with hydrocarbons. The data (Table 1) demonstrated that oil removal by natural attenuation without nutrients supplementation or bacteria addition was much slower compared to bioaugmentation. After 28 days, only about 38% of crude oil concentration of 0.5% (w/v) was removed by natural attenuation. Addition of Solid waste dates and corn steep liquor showed a significant acceleration of crude oil degradation. In fact, the experimental results showed that the stationary phase has excellent selectivity and also possesses a good recognition ability toward these organic compounds, especially solid waste dates and as shown in Figure 4. |
| Aneja [34] reported that biosurfactant can be synthesized from low cost substrates, such as molasses and corn steep liquor, for use in environmental applications. The addition of both the biosurfactant and/ or bacterial cells of P. cepacia favored the biodegradation of hydrophobic organic compounds. It is evident to the results that the biosurfactant alone, and its producer species are both capable of stimulating biodegradation to a large extent. Thus, Silva EJ et al. and Nikolopoulou et al. [35,36] have reported that identifying the key organisms that play an important role in different bioremediation treatments for understanding, evaluating and further decide upon the best in situ bioremediation strategy. To reduce medium costs of date's molasses, and corn steep liquor (CSL), a by-product of wet milling corn for starch, are extensively used in bioremediation as a component of microorganism culture medium. It provides a rich but economical source of nutrients, rich sugar, amino acids, organic acids, vitamins, and minerals [37,38]. |
| Even though the process is site specific to Libya, however, aspects of isolation process, DNA identification and biodegradation experimental methods can also be applied to other similar hydrocarbons properties elsewhere. |
| Conclusion |
| Bacteria strain SAR1 was isolated and characterized belonging to the genus Kocuria according to its 16S rDNA, as well as biochemical characteristics. DNA–DNA relatedness indicates that strain SAR1 is a member of the same genomic species. It is Gram-positive, large, opaque, shiny colonies with serrated edges, occurred in pairs and sometimes in chains or in a group, aerobic, oxidase-negative, catalasepositive. Kocuria sp. strain SAR1 can utilize crude oil hydrocarbons as the sole carbon source. Strain SAR1 has a high ability to degrade crude oil in a pH 7, temperature 30 C°. The degradation efficiencies and the result showed that the isolate could remove 68%, 70% in 28 days when cultivated with 0.2% (w/v) of solid waste dates and corn steep liquor, respectively. The use of pretreated solid waste dates which forms a byproduct of the agro- industry as the state of Libya has an easy access to enhance the bioremediation of oil spills, may form a useful method to be applied. |
| Acknowledgment |
| We thank Mr. Youssef Imrimi, director, Maintenance Department (AGOCO) for his encouragement and support. The Arabian Gulf Oil Co. of Libya funded this work. |
References
|
Tables and Figures at a glance
| Table 1 |
Figures at a glance
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 16777
- [From(publication date):
July-2015 - Jul 05, 2025] - Breakdown by view type
- HTML page views : 12048
- PDF downloads : 4729
