Performance Evaluation of Linseed (Linum usitatissimum L.) Varieties in Buno Bedele Zone, South Western Oromia, Ethiopia
Received: 01-Jul-2024 / Manuscript No. acst-24-145091 / Editor assigned: 04-Jul-2024 / PreQC No. acst-24-145091 / Reviewed: 18-Jul-2024 / QC No. acst-24-145091 / Revised: 22-Jul-2024 / Manuscript No. acst-24-145091 / Published Date: 29-Jul-2024
Abstract
This experiment was carried out using twelve linseed varieties namely;(Bekoji-14, Kassa-2, Welen, Bekoji, Kuma, Yadano, Furtu, Bakalcha, Dibane, Horesoba, Jitu and local check) were sown in RCBD with three replications during the 2022 and 2023 main cropping season with the objective, to evaluate the performance of improved linseed varieties and their genetic variability for seed yield and related traits in to study areas. All important data were collected and analyzed by using R-software accordingly. Combined analysis of data from the three locations revealed that there is significant difference among varieties for days to flowering, days to maturity and grain yield, but non-significant for plant height (cm), number of primary branches per plant and number of capsules per plant. Significant effect of location was observed in plant height, number of primary branch per plant, number of capsule per plant and grain yield however non-significant in days to flowering and days to maturity. The interaction of Variety X location was significant for days to flowering, days to maturity, plant height and grain yield, however non-significant for number of primary branch per plant and number of capsule per plant. The maximum seed yield was recorded in variety Kuma (1588.9 kgha-1) followed by Beokoji-14 (1476.7 kgha-1) and the lowest yield (978.9 kgha-1) was obtained from local check. The combined AMMI analysis for seed yield across environments revealed significantly affected by environments that hold 26.6% of the total variation. The genotype and genotype by environmental interaction were significant and accounted for 12.40% and 19.42% respectively. Principal component 1 and 2 accounted for 10.78 % and 6.13% of the GEI respectively with a total of 16.91% variation. Therefore, Kuma and Bekoji-14 varieties were identified as the best varieties for yielding ability, stability and recommended in the area and with similar agro-ecologies.
keywords
Evaluation; Linseed; RCBD, AMMI; Varieties; Interactions; Stability
Introductions
Among oil crops linseed (Linum usitatissimum L.) is one of the ancient oilseeds cultivated for oil, food and fiber (Nozkova et al., 2016). The haploid chromosome number of Linseed is 15 (n =15), and the diploid chromosome number is 30 (2n =30). It is an important oil crop cultivated worldwide for oil and fiber. Ethiopia’s oilseed crop, which is quickly expanding to fulfill both domestic and international demand, is critical to the country’s foreign exchange revenues and income. Oilseed exports account for about 11.5% of Ethiopia’s total export profits. Oilseeds supplied 5.90% (766,167.66 ha) of grain crop area and 2.27% (7,774,444.17 quintals) of grain production to Ethiopia’s national grain total (CSA, 2021). Niger seed, sesame, and linseed accounted for 1.48% (19.766.00 ha), 2.85% (369,897.32 ha), and 0.61% (78,921.37 ha), respectively, of the grain crop area. These crops account for 0.63%, 0.76%, and 0.24% of grain crop production, respectively. The European top producer is the Russian Federation (13.86%) (Jovovic et al.,2019). The crop is largely grown in temperate climates and cool tropics, including the highlands (>2500 m above sea level) of Ethiopia. It thrives best at altitudes of 2200–2800 m above sea level in Ethiopia, but it can also be grown at 1200 m and 3420 m above sea level (Behailu et al., 2019). Linseed grows in cold weather ranging from 10–30°C, but it produces the best harvests when temperatures are between 21–22°C during the growth period. In fact, linseed is the second biggest oil plant in terms of acreage cultivated and yield in the highland areas of Ethiopia, after Niger seed (W. Adugna ., 2000) [1]. In Ethiopia, linseed supplies 10.3% (78,921.37 ha) of the oilseed crop area and 10.35% (80,456.64 tons) of the production of the national oilseed total (CSA, 2021). Despite its diverse uses for the local oil sector and foreign currency earnings, linseed productivity in Ethiopia is characterized by low yield, with an average yield of 10.2 qt ha-1 (CSA, 2021) compared to more than 1.5 t/ha in developed countries (FAO, 2008). In Oromia region linseed covered 27,661.60 hectares of cultivated land and has a production of about 310,824.57 quintals with average yield of 11.24 qt ha-1 (CSA, 2021). This indicates in the Oromia region linseed production potential is better. Even though there is no production status of the study area for this crop appropriate improved variety is crucial in order to increase the production.
Some attempts have been done to improve the limitation of ideal linseed varieties for the area of Buno Bedele zone by Jimma Agricultural Research Center (JARC). However, that is not enough and satisfying the farmers demand regarding the option and accessibility of improved varieties. Hence, this study was conceived to evaluate the newly released linseed varieties to the agro-ecology of the Buno Bedele zones of south western Oromia region, Ethiopia and identify the most high yielders and stable varieties for the farmers. One of the major production barriers affecting linseed productivity is lack of access to improved varieties, which is why the majority (>90%) of Ethiopian farmers use unimproved seeds (FAO, 2010) [2]. On the contrary, the Ethiopian Institute of Agricultural Research has released more improved linseed varieties (MoA, 2016) that, if properly assessed and produced by farmers, have the potential to increase crop output. Every crop improvement effort begins with a breeder looking into the existence of genetic variability for the desired traits (Kahsay et al., 2020). Ethiopia is believed to be the secondary place of variation, and it is the world’s fifth largest linseed producer (W. Adunya, 2007). Ethiopia’s diverse agro climatic conditions may have led to the country’s linseed crop diversification. However, having variance in a population is not enough to improve desired qualities and little or no research on the genetic variability of released linseed varieties has been conducted. As a result, breeders must evaluate the degree and distribution of genetic diversity in the genetic materials that are readily available [3]. The crop genotypes grown in different environments would frequently encounter significant fluctuations in yield performance, particularly when the growing environments are distinctly different, the test genotypes differentially respond to changes in the growing environments or both. AMMI is a multivariate tool, which was highly effective for the analysis of multi environment trials and in the recent years, this method has often been used by international agricultural development agencies (Grüneberg et al., 2005). The most recent method, the GGE (genotype main effect (G) plus G x E interaction) biplot model, provides breeders a more complete and visual evaluation of all aspects of the data by creating a biplot that simultaneously represents mean performance and stability, as well as identifying mega-environments (Ding et al., 2007; Yan and Kang, 2003). In this study, we attempted to apply AMMI and sites regression GGE biplot statistical model for determination of the magnitude and pattern of G × E interaction effects and performance stability of seed yield of 12 linseed genotypes [4].
Materials and Methods
Description of the study Areas
The field experiment was conducted during the 2022 – 2023 G.C main cropping seasons for two years at three districts (Bedele, Dabo Hana and Didesa) of Buno Bedele Zone, South Western Oromia where agro ecology assumed to be conducive for Linseed production (Table 1).
| Geographical Coordinates | Ave. Temperature (OC) | ||||||
|---|---|---|---|---|---|---|---|
| Locations | Altitude(m.a.s.l) | R/fall (mm) | Soil Type | Latitude N | Longitude E | Max. | Min. |
| Bedele | 1300-2200 | 1200-1800 | Nitosols | 8°28'60.00" | 36°20'60.00" | 28.5 | 12.5 |
| Dabo Hana | 1791-1990 | 1300-1945 | Nitosols | 8°55ʹ60 20” | 36°26ʹ 19.00” | 25.8 | 12.9 |
| Didessa | 1945-2340 | 1250-2000 | Nitosols | 8°04'60.00" | 36°39'59.99" | 28 | 13 |
| Source: Different literatures | |||||||
Table 1: Agro-ecological features of the experimental Locations.
Experimental materials
Eleven improved linseed varieties from the Holetta, Kulumsa and Sinana Agricultural Research Centers, as well as one local check from farmers, were collected and evaluated for performance and variability over seasons (Table 2).
Varieties |
Year of release | Releasing center/Maintainer | Seed Color |
|---|---|---|---|
| Bekoji-14 | 2014 | HARC (EIAR) | Brown |
| Kassa-2 | 2012 | HARC (EIAR) | Yellow |
| Welen | 2019 | KARC (EIAR) | Brown |
| Bekoji | - | SARC (EIAR) | Brown |
| Kuma | 2016 | KARC (EIAR) | Brown |
| Yadano | 2015 | KARC (EIAR) | Brown |
| Furtu | 2013 | KARC (EIAR) | Brown |
| Bakalcha | 2010 | KARC (EIAR) | Brown |
| Dibanne | 2009 | SARC (OARI) | Brown |
| Horesoba | 2019 | SARC (OARI) | Brown |
| Jitu | 2012 | SARC (OARI) | Brown |
| Local | - | - | Brown |
| Remark: HARC=Holetta Agricultural Research Center, KARC=Kulumsa Agricultural Research Center, SARC=Sinana Agricultural Research Center, EIAR=Ethiopia Institute of Agricultural Research & OARI=Oromia Agricultural Research Institute | |||
Table 2: Description of the linseed varieties used for this study.
Experimental design and field work
The treatments were laid with a randomized completed block design (RCBD) with three replications was used in the study. The experimental plots were 1.5 m × 2 m (3m2) in size, with rows separated by 0.3m. The distance between plots and blocks was 0.5 m and 1m, respectively. Each plot had five rows, with the middle three used for the collection of the data and the two outermost rows were used as border rows. The experimental sites were plowed three times with draught animals called oxen before planting, and 25 kg ha−1 seed rate was used for planting. After weighing the seeds with a sensitive balance, they were assigned to each row, and drilling was used for planting. Planting occurred during 2022 and 2023 main cropping seasons, following the onset of rain and when the locations received the moisture required for germination. Hand weeding was used three times during the experiment’s growing seasons, and 30 kg ha−1 of urea and 50 kg ha−1 of NPS fertilizer were used according to crop recommendations. Harvesting begins in the last week of November of each cropping year, based on the maturity time of the varieties [5].
Data Collection
Data on flowering days, maturity days, number of primary branch/plant, capsule number/plant, plant height, and seed yield per plot were collected from the three middle rows. Data for days to flowering, days to maturity, and seed yield were collected on a plot basis on three middle rows.
Days to flowering - were calculated from the date of planting when 75% of the crop stand produced the first flower.
Days to maturity - The number of days from planting to physiological maturity of the plants was used to compute the days to maturity.
Plant height - the average height of five randomly selected plants, measured from the base to the tip of the plant.
The number of primary branch/plant - was recorded as the average number of primary branch from five randomly sampled plants taken from the three middle rows of the plots
The number of capsules per plant - It was calculated as the mean number of capsules collected from five randomly selected from the three middle rows of the plots.
Seed yield (kg plot−1) - It was calculated as the entire seed yield produced from the plants harvested and threshed and converted into seed yield per hectare [6].
Analysis of variance
The ANOVA was run for the three locations separately and combined over the three locations for all characters using R software. Analysis of variance was carried out to partition the variance due to genotype, environment and genotype by environment interaction, replication within environment and block within replication. The combined analysis of variance was carried out to estimate the additive effects of the environment, genotype and GEI. Mean separations were estimated using Least Significant Difference (LSD) for the comparison among the experimental varieties at 0.05 probability level.
ANOVA model for a single location as: Yijk= µ + Gi +Rj + Bkj + Eijk
ANOVA model for the combined over locations as: Yijkr = µ +Gi + Lj + Rk(Lj) + Br(LjRk) + GiLj + Eijkr., Where Y is the performance of genotype i in jth location, kth replication and rth incomplete block, μ is the mean effect, Lj is the effect of jth location, Rk(Lj) the effect of the kth replication in location J, Br (LjRk) is the effect of rth incomplete block in jth location and kth replication, GiLj is the interaction effect of genotype i and location j and εijkr is the error associated with the ith genotype, jth location, kth replication, rth incomplete block
Stability analysis
Yield stability of the genotypes was evaluated using different models: Yield stability index (YSI), AMMI Stability Value (ASV), Additive Main Effect and Multiplicative Interaction (AMMI) model and Genotype Main Effect and Genotype x Environment Interaction Effect (GGE) biplot analysis were used to determine the effects of GEI on yields. These analyses also were performed using R software [7].
Yield stability index (YSI)
This measurement was developed by (Farshadfar et al., 2011). Stability by itself should not be the only measurement for selection, because the most stable genotypes would not necessarily give the best yield performance. This method is vital to measure and rank genotypes based on grain yield stability. The summation of rank of ASV and rank of yield are used to calculate YSI. The genotype with least YSI is considered as the most stable with high grain yield (Dabesa et al., 2016).
YSI was calculated as: YSI = RASV + RY, Where: RASV is the rank of AMMI stability value, RY is the rank of mean yield of genotypes across environments.
AMMI stability value
The ASV is the distance from the coordinate point to the origin in a two dimensional scatter length of IPCA1 scores against IPCA2 scores in the AMMI model (Purchase, 1997). AMMI Stability Value (ASV), length of genotype and environment markers of the origin in a two dimensional plot of IPCA1 scores against IPCA2 scores was calculated as follow (Purchase, 1997).
Where: IPCA1= interaction principal component axis 1; IPCA2 = interaction principal component, axis 2. Genotypes with lower values of the ASV are considered to be more stable
Additive main effect and multiplicative interaction (AMMI) model
Additive Main Effect and Multiplicative Interaction (AMMI) is one of most widely used model to explain G×E interaction of multi-environment genotype trial and categorizing the genotypes into narrow or wider adaptation (Crossa et al., 1990). The AMMI analysis uses analysis of variance (ANOVA) followed by a principal component analysis applied to the sums of squares allocated by the ANOVA to the GEI (Kempton et al., 1984).
The AMMI Model Equation is: Ῡijk =µ+Gi +Ej +Σmk=1 λkαikγjk +Рij
Where: Ῡijk. = the yield of the ith genotype in the jth environment, Gi = the mean of the ith genotype minus the grand mean, Ej = the mean of the jth environment minus the grand mean, λk = the square root of the Eigen value of the kth IPCA axis, αik and γjk = the principal component scores for IPCA axis k of the ith genotypes and the jth environment, Рij = the deviation from the model [8].
Genotype main effect and genotype by environment interaction (GGE) biplot
The GGE biplot (Purchase, 1997) model is: Yij -μ-βj = λ1Ԑi1ηj1 + λ2Ԑi2ηj2 + Ԑij
where: Yij= the performance of the ith genotype in the jth environment; μ = the grand mean; βj = the main effect of the environment j: λl and λ2 = Singular value for IPCA1 and IPCA2 respectively: Ԑi1 and Ԑi2 = Eigen vectors of genotype i IPCA1 and IPCA2 respectively: ηj1 and ηj2 = Eigen vectors of environment j for IPCA1 and IPCA2 respectively; Ԑij = Residual associated with genotype i and environment j.
Results and Discussions
Analysis of variance
The ANOVA revealed highly significant differences (p<0.01) for seed yield at Bedeele-2023 only and significance differences (p<0.05) at Dabo Hana-2022, Dabo hana-2023, Bedele-2022 and Didesa-2023 [9].
Seed yield
The average environmental seed yield across varieties ranged from the lowest of 1088.7 kgha-1 at Didesa -2023 to the highest of 1662.9 kgha-1 at Dabo Hana-2023), with a grand mean of 1294.6 kgha-1. The varieties average seed yield across environments ranged from the lowest of 978.9 kgha-1 for local check to the highest of 1588.9a kgha-1 for Kuma. This difference could be due to their genetic potential of the varieties, and also environment explained large variation indicated the existence of diverse mega environments (Table 3).
| Mean of seed yield of 12 linseed varieties across environment | |||||||
|---|---|---|---|---|---|---|---|
| Varieties | D/Hana-2022 | D/Hana-2023 | Bedele-2022 | Bedele-2023 | Didesa-2023 | Mean | YAd.(%) |
| Bekoji 14 | 2288.89a | 1400.00bc | 1233.33abc | 1444.4a | 1016.7bc | 1476.7ab | 50.85 |
| Kassa-2 | 1633.33bcd | 1434.44abc | 1177.78bc | 1077.8abc | 1000.0bc | 1260.2bc | 28.74 |
| Welen | 1511.11bcd | 1544.44ab | 1144.44bc | 1400.0a | 1066.7bc | 1333.3abc | 36.2 |
| Bekoji | 1633.33bcd | 1355.56bcd | 1033.33c | 773.3cd | 1225.6ab | 1204.2cd | 23.02 |
| Kuma | 1788.89b | 1527.78abc | 1566.67a | 1627.8a | 1433.3a | 1588.9a | 62.31 |
| Yadano | 1322.22cd | 1722.22a | 1077.78c | 1244.4abc | 1277.8ab | 1328.9bc | 35.75 |
| Furtu | 1755.56bc | 1416.67bc | 1455.56ab | 788.9bcd | 1233.3ab | 1330.0abc | 35.87 |
| Bakalcha | 1577.78bcd | 1090.00d | 1146.67bc | 1264.4abc | 794.4c | 1174.7cd | 20 |
| Dibanne | 1744.44bcd | 1473.33abc | 1077.78c | 1466.7a | 983.3bc | 1349.1abc | 37.82 |
| Horesoba | 1725.00bcd | 1296.67bcd | 1250.00abc | 1373.3ab | 1058.3bc | 1319.3bc | 34.77 |
| Jitu | 1750.00bcd | 1368.33bcd | 1000.00c | 694.4cd | 950.0bc | 1191.3cd | 21.7 |
| Local | 1255.56d | 1244.44cd | 1066.67c | 338.9d | 988.9bc | 978.9d | - |
| Mean | 1662.96 | 1404.17 | 1192.78 | 1124.5 | 1088.7 | 1294.6 | |
| LSD (0.05) | 440.23 | 291.8 | 378.56 | 590.61 | 322.2 | 259.8 | |
| CV % | 16.08 | 12.16 | 17.87 | 31.02 | 16.95 | 27.8 | |
| P-value | * | * | * | ** | * | ** | |
| Remark: LSD= Least significant different, CV= Coefficient of variation, *=significant at P<0.05 level, **=highly significant YAd.=Yield Advantage Over local check, | |||||||
Table 3: Mean seed yield (kgha-1) of twelve (12) linseed varieties evaluated at five environments.
Yield related traits
The results of the combined analysis of variance are depicted. The combined analysis of variance over the five sites for two seasons revealed a significant (P < 0.01) difference in flowering days and maturity days however, non-significant for plant height, number of primary branch and number of capsule per plant between the studied varieties. Accordingly, the tested linseed varieties shows statistically significant variation that ranged from 47.1(Kassa-2) and 85.0 (Local check) for days to flowering and 127.8 (Kassa-2) to 153.4 (local check) for days to maturity. From this result kassa-2 was the earliest to maturity whereas local check was the late maturing variety. A similar result was reported by (F.Amsalu et al., 2020) (Table 4).
| Varieties | DTF (days) | DTM (days) | PLH (cm) | NPB/PL(no) | NC/PL(no) | Disease Reaction |
|---|---|---|---|---|---|---|
| Bekoji-14 | 59.2cd | 144.9b | 80.4 | 5.2 | 27.6 | 0 |
| Kassa-2 | 47.1g | 127.8f | 81.2 | 5.7 | 27.9 | 0 |
| Welen | 52.9f | 131.5e | 74.7 | 4.4 | 23.8 | 0 |
| Bekoji | 56.9de | 142.9bcd | 80.1 | 4.9 | 26.9 | 0 |
| Kuma | 62.8bc | 142.0d | 79.4 | 5.2 | 27.3 | 0 |
| Yadano | 56.9de | 144.3bc | 70.9 | 4.2 | 23.3 | 0 |
| Furtu | 63.7b | 131.7e | 77.9 | 4.4 | 22.1 | 0 |
| Bakalcha | 54.0ef | 130.0e | 73.8 | 4.4 | 21.6 | 0 |
| Dibanne | 57.3de | 142.8bcd | 78.3 | 4.0 | 23.8 | 0 |
| Horesoba | 57.2de | 142.2cd | 76.5 | 4.6 | 23.5 | 0 |
| Jitu | 56.6de | 142.6cd | 79.3 | 4.4 | 25.1 | 0 |
| Local | 85.0a | 153.4a | 78.4 | 4.5 | 22.7 | 0 |
| Grand Mean | 59.1 | 139.68 | 77.6 | 4.7 | 24.6 | |
| LSD (0.05) | 3.6 | 2.15 | 6.9 | 1.0 | 6.2 | |
| CV % | 8.5 | 2.14 | 12.3 | 9.6 | 14.7 | |
| P-value | *** | *** | NS | NS | NS | |
| Remark: DTF= Days to Flowering, DTM= Days to Maturity, PLH= Plant height (cm), NPB/PL= Number of primary branch per Plant, NC/PL= Number of capsule per Plant, LSD= Least significant different, CV= Coefficient of variation, NS= Non-significant, *** significant at 0.001 probability level | ||||||
Table 4: Combined mean yield and other parameters of linseed variety adaptation trial across the three locations.
Combined Analysis of Variance for seed yield and yield-related traits
The combined analysis of variance showed that significant differences were recorded across location for most of parameters except number of primary branch per plant and number of capsule per plant. After testing the homogeneity of error variances of the test locations, the combined analysis of variance was conducted for each trait with special focus on grain yield and other agronomic traits in order to examine the presence of significant effect of locations, varieties and varieties x location interactions [10].
The mean square of analysis of variance (ANOVA) is presented. Significant differences were detected among the main and the interaction effects at (p ≤ 0.05) and (p<0.001) for most of the parameters (Table 5).
SOV |
DF | DTF(days) | DTM(days) | PLH(cm) | NPB/PL(no) | NC/PL(no) | SY (kgha-1) |
|---|---|---|---|---|---|---|---|
| Loc. | 2 | 11.07ns | 9.20ns | 1947.41*** | 15.22*** | 1222.38** | 2064407** |
| Rep. | 2 | 0.71ns | 21.05* | 37.44ns | 0.3098ns | 26.87ns | 3895ns |
| Yr. | 1 | 18.78ns | 1.00ns | 2557.94*** | 56.94*** | 1047.82*** | 962579*** |
| Var. | 11 | 1278.28*** | 877.47*** | 141.46*** | 2.86* | 76.42ns | 356370** |
| Var.*Loc. | 22 | 151.33*** | 28.26*** | 69.06** | 2.17ns | 51.63ns | 104936* |
| Var.*yr. | 11 | 4.54ns | 10.44* | 90.67* | 1.35ns | 71.10ns | 166036** |
| Loc.*yr. | 2 | 2.35ns | 3.55ns | 40.30ns | 0.04ns | 1205*** | 163402* |
| Var.*Loc.*yr. | 22 | 1.17ns | 3.37ns | 13.10ns | 0.89ns | 18.51ns | 182392** |
| Pooled error | 118 | 6.19 | 5.42 | 38.56 | 1.52 | 41.73 | 60645 |
| CV | 8.5 | 2.14 | 12.3 | 9.6 | 14.7 | 27.8 | |
| Remark: SOV=source of variations, DF=degree of freedom, DTF=days to flowering, DTM=days to maturity, PLH=plant height, NPB/PL=number of primary branch per plant, NC/PL=number of capsule per plant, GY=grain yield, Loc.=Location, Rep=replication, yr.=year, Var.=variety, CV=coefficient of variations, *=significant 0.05 probability level, **=significant at 0.01 probability level, ***=significant at 0.001 probability level | |||||||
Table 5: Mean square value of combined Analysis of Variance (ANOVA) for seed yield and yield related traits of twelve (12) Linseed varieties tested at three locations for two years.
Stability analysis
Yield stability index (YSI)
Yield stability index incorporates both mean yield and stability in a single criterion. The minimum values of YSI desirable genotypes with high mean yield and stability.
AMMI stability value (ASV)
The ASV measure was proposed to cope up the fact that the AMMI model does not make a provision for a quantitative stability measure (Purchase, 2000). Depending on this method, genotype with least ASV score is the stable, accordingly, variety Kassa-2 followed by Kuma and Horesoba were the most stable respectively. While local, Bekoji-4, Furtu, Dibanne and Jitu were unstable varieties. The greater the IPCA scores (Negative or Positive), the more specifically adapted variety is to certain environment. The closer the IPCA scores to zero, the more stable the genotype over the tested locations. The further away from zero the IPCA score for the environments is the more interaction the environment has with the genotypes, thus making difficult to choose genotypes for that environment (Table 6) [11].
| Varieties | Yield (kgha-1) | rY | PCA1 | PCA2 | ASV | rASV | YSI |
|---|---|---|---|---|---|---|---|
| Bekoji-14 | 1476.7 | 2 | 9.72 | -16.4 | 23.69 | 11 | 13 |
| Kassa-2 | 1260.2 | 8 | -0.29 | 0.68 | 0.85 | 1 | 9 |
| Welen | 1333.3 | 4 | 6.44 | 8.88 | 14.38 | 4 | 8 |
| Bekoji | 1204.2 | 9 | -9.45 | -1.37 | 16.67 | 5 | 14 |
| Kuma | 1588.9 | 1 | 5.04 | 5.01 | 10.74 | 2 | 3 |
| Yadano | 1328.9 | 6 | -1.48 | 17.55 | 17.74 | 6 | 12 |
| Furtu | 1330 | 5 | -12.22 | -5.04 | 22.06 | 10 | 15 |
| Bakalcha | 1174.7 | 11 | 10.04 | -2.98 | 17.9 | 7 | 18 |
| Dibanne | 1349.1 | 3 | 10.5 | 1.01 | 18.47 | 9 | 12 |
| Horesoba | 1319.3 | 7 | 7.85 | -0.22 | 13.8 | 3 | 10 |
| Jitu | 1191.3 | 10 | -9.02 | -9.02 | 18.25 | 8 | 18 |
| Local | 978.9 | 12 | -17.49 | 1.91 | 30.79 | 12 | 24 |
| Remark: rY=rank in yield, IPCA=interaction principal component, ASV=ammi stability value, rASV = rank in ammi stability value, YSI=yield stability index | |||||||
Table 6: The first and second IPCA, Grain Mean yield and different yield stability statistics investigated in linseed varieties over locations.
Additive main effects and multiple interaction (AMMI) model
The mean squares for all varieties evaluated under different environmental condition for seed yield are presented below. The result indicated that differences among all varieties were significant (P ≤ 0.01). Variation due to genotypes by environments interaction was significant for the studied traits, indicated that genotypes differ genetically in their response to different environment. The genotypes by environments interaction was significant effect on the seed yield, which explained 19.42% of the total variation whiles the genotypes, contributed 12.40% of the variation. However, large portion (26.11%) of the total variation was attributed to the environmental effect. The environment explains a major amount of the yield differential, showing that the environments were different, according to AMMI (Table 7) [12].
Source |
D.F. | S.S. | EX.SS % | M.S. |
|---|---|---|---|---|
| Environments | 4 | 8E+06 | 26.11 | 2064407** |
| Genotypes | 11 | 4E+06 | 12.4 | 356370* |
| Interactions (G x E) | 44 | 6E+06 | 19.42 | 139575** |
| PCA1 | 14 | 3E+06 | 10.78 | 243477* |
| PCA2 | 12 | 2E+06 | 6.13 | 161623* |
| PCA3 | 10 | 642666 | 2.03 | 64267ns |
| PCA4 | 8 | 150494 | 0.48 | 18812ns |
| Residuals | 110 | 7E+06 | 21.17 | 60876 |
| Total | 223 | 3E+07 | 100 | 141813 |
| Remark: DF=degree of freedom, SS=sum of square, EXSS=explained sum of square, MS=mean sum of square, PCA= = Interaction Principal Component Axis | ||||
Table 7: Additive main effect and multiplicative interaction analysis of variances (AMMI) for seed yield of twelve Linseed varieties.
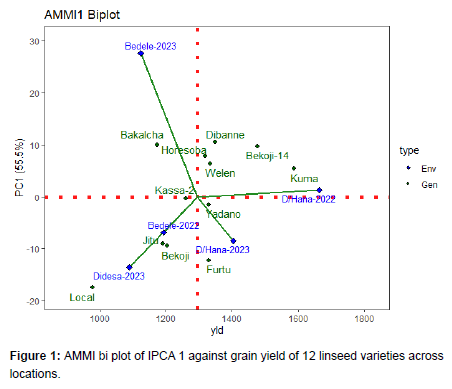
AMMI 1 biplot
In AMMI biplot 1 showing main effects means on the abscissa and principal component (IPCA) values as the ordinates, genotypes (environments) that appear almost on a perpendicular line have similar means and those that fall on the almost horizontal line have similar interaction patterns. Genotypes that group together have similar adaptation while environments which group together influences the genotypes in the same way. Genotypes (environments) with large IPCA1 scores (either positive or negative) have high interactions whereas genotypes (environments) with IPCA1 score near zero have small interactions.
AMMI-1 considers genotype and environment main effects plus the PCA1 to interpret the residual matrix and represented genotype productivity. It is further stated that any genotype with PCA1 value close to zero shows general adaptation to the tested locations whereas a large genotypic PCA1 score reflects more specific adaptation to location with PCA1 scores of the same size. Genotypes and environments with IPCA1 scores of the same sign produce positive interaction suggesting adaptation of genotypes in those locations whereas the reverse sign of PCA value of genotypes and locations depicts negative interaction i.e., poor performance of genotypes in such locations. In summary, a stable genotype might not be the highest yielding. Genotypes having a zero IPCA1 score are less influenced by the locations and adapted to all locations [13].
The closer the IPCA score to zero, the more stable the genotypes over the tested environments. Since IPCA1 scores of Linseed varieties Welen, Horesoba and Kassa-2, were close to zero, they were more stable varieties that across these locations. However, the mean yield of Kassa-2 had a mean yield below average, therefore, they are least preferable. A variety showing high positive interaction in environments has the ability to exploit the agro ecological and agro-management conditions of the specific location and is therefore best suited to that location. In this case, Kuma and Bekoji-14 linseed varieties are suited for Dabo Hana. Linseed varieties Furtu is suited for Dabo Hana-2023. Linseed varieties Bakalcha is suited for Bedele-2023 and Jitu variety is suited for Bedele-2022 (Figure 1).
GGE Bi-plot analysis for grain yield
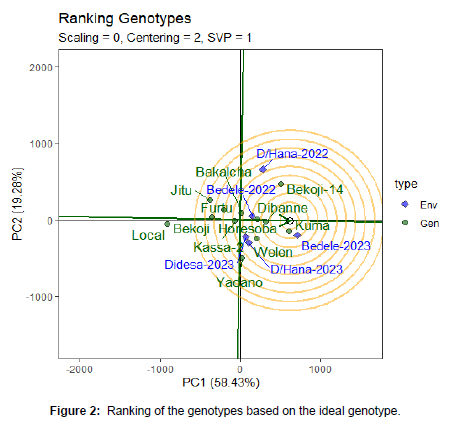
Ranking of Genotypes
Stability can be identified using concentric circles and also ideal genotypes are at the center of the concentric circle. The ideal genotype is the one that with the highest mean performance and absolutely stable (Purchase, 2000). The genotypes that are closer to the ideal genotypes are the best performing genotypes. Hence, the GGE bi plots shows that Kuma is an ideal variety, with other varieties, like Dibane, Horesoba, Welen and Bekoji-14 are desirable varieties as they are closer to the ideal variety on the bi plot. The varieties local and Jitu are the most undesirable varieties as they are too far to the ideal variety on the bi plot (Figure 2).
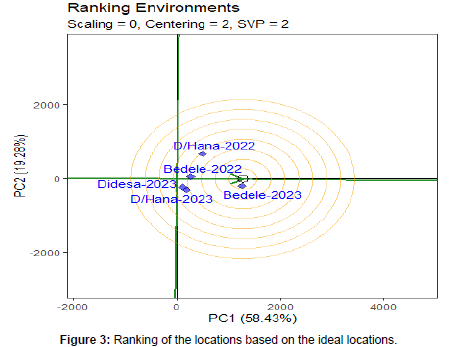
Ranking of environments
The most representative of the locations (ability to represent the mega environment) and the most powerful to discriminate varieties reported that the ideal environment is the one located at the center of the concentric circles, and it is possible to identify desirable environments based on their closeness to the ideal environment (Rad et al., 2013) reported that a testing location has less power to discriminate genotypes when located far away from the center of the concentric circle or to an ideal location. Therefore, among the test locations, location Bedele which fell into the center of concentric circles was an ideal test location in terms of being the most representative of the overall locations and the most powerful to discriminate the performance of the tested genotypes. Next to the first concentric circle location, locations Dabo Hana-2022 is close to the ideal location while, Didesa is detected as the weakest locations to discriminate varieties (Figure 3).
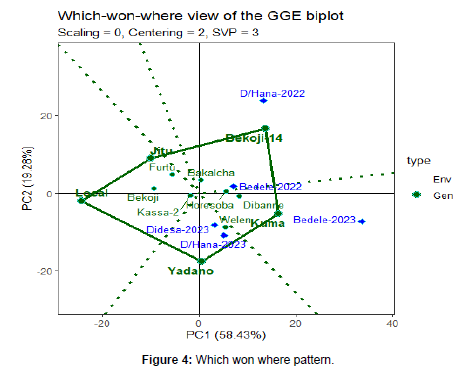
Which-won-where pattern
The polygon view of GGE bi plot indicates the best genotypes in each environment and group of environments (Yan et al., 2002). In this situation, the polygon is formed by connecting the genotypes that are farthest away from the bi plot origin, such that all the other genotypes are contained in the polygon. In this case, the polygon connects all the farthest genotypes and perpendicular lines divide the polygon into sectors. Sectors help to visualize the mega-environments.
This means that winning genotypes for each sector are placed at the vertex. Polygon view of the linseed varieties tested at three locations presented in. The genotypes found at the vertex of the polygon perform best in the environments within the sector (Tinker et al., 2006). Six rays divide the bi plot in to six sector and the locations fall in to three different mega-environments.
Accordingly, Yadano, Kuma, Bekoji-14, Jitu and local varieties were the vertex varieties. From this figure, Yadono variety best performer at Didesa and Dabo Hana 2023 in mega environment. The second environment containing the higher yielding environment Bedele-2023, with winner variety Kuma. The third environment includes Dabo Hana-2022 with a winner variety Bekoji-14. From the figure, Jitu and local had no environment on the vertex. This indicates that varieties in the vertex without environment performed poorly in all the locations (Figure.4) [14].
Conclusion and Recommendation
Combined analysis of variance (ANOVA) result revealed significant difference of seed yield and most of yield contributing traits among evaluated Linseed varieties across locations, years and the interactions. This indicated that, the location and fluctuation of weather condition over the cropping season had affected performance of varieties. Although the GEI of grain yield partitioned in to different IPCAs using AMMI model analysis, the first principal component axis for interaction alone explains most of the interaction sum of squares. The sign and magnitude of IPCA scores showed the relative contribution of each genotype and environment for the genotype and environment interactions.
This helps to summarize the pattern and magnitude of GEI and main effects that reveal clear insight into the adaptation of genotypes to environments. This shows that, Kuma and Bekoji-14 varieties are fewer contributors to the interaction effect and have consistent performances across locations. Therefore, Kuma and Bekoji-14 were identified as the best varieties in terms of yielding ability, stability and better agronomic performance.
Acknowledgments
The authors greatly acknowledged Oromia Agricultural Research Institute (IQQO) for the financial support and Sinana, Kulumsa and Holetta Agricultural Research Centers are also highly acknowledged for providing the linseed improved varieties also all research staffs for technical support.
References
- Behailu S, Yechalew H, Mesfn A, Abush T (2019) Linseed (Linum usitatissimum L.) Variety Adaptation at South Western Ethiopia 5: 41-45.
- Crossa J, Gauch Jr HG, Zobel RW (1990) Additive main effects and multiplicative interaction analysis of two international maize cultivar trials. Crop Science 30: 493-500.
- Dabessa A, Alemu B, Abebe Z, Lule D (2016) Genotype by Environment Interaction and Kernel Yield Stability of Groundnut (Arachis hypogaea L.) Varieties in Western Oromia, Ethiopia. Journal of Agriculture and Crops 2:113-120.
- Farshadfar E, Zali H, Mohammadi R (2011) Evaluation of phenotypic stability chickpea genotypes using GGE-Biplot. Ann Biol Res 2: 282-292.
- Amsalu F (2020) Estimates of heritability, genetic and principal components analysis for yield and its traits in linseed genotypes (Linum usitatissimum L.) in central highlands of Ethiopia. International Journal of Research in Agriculture and Forestry7:1-6.
- Grüneberg WJ, Manrique K, Zhang D, Hermann M (2005) Genotype × Envinronment Interactions for a Diverse Set of Sweetpotato Clones Evaluated across Varying Ecogeographic Conditions in Peru. Crop Science 45: 2160-2171.
- Kahsay AG, Birhanu K (2020) Yield evaluation and character association of linseed (Linum usitatissimum L.) genotypes in moisture stress areas of South Tigray, Ethiopia. Journal of Cereals and Oilseeds 11: 16-20.
- Kempton RA (1984) The use of biplots in interpreting variety by environment interactions. The Journal of Agricultural Science 103: 123-135.
- Nozkova J, Martin P, Marie V, Nina B, Eva T, et al. (2016) Descriptor list of flax (Linum usitatissimum L.). Slovak University of Agriculture ISBN978-80-552-1484-9.
- Purchase JL, Hatting H, Deventer CS van (2000) Genotype × environment interaction of winter wheat (Triticum aestivum L.) in South Africa: II. Stability analysis of yield performance. South African Journal of Plant and Soil 17: 101-107.
- Purchase H (1997) Which aesthetic has the greatest effect on human understanding? In International Symposium on Graph Drawing (pp.248-261).
- Rad MR Naroui (2013) Genotype environment interaction by AMMI and GGE biplot analysis in three consecutive generations of wheat (Triticum aestivum) under normal and drought stress conditions. Australian Journal of Crop Science 7.7: 956.
- Tinker NA, Yan W (2006) Information systems for crop performance data. Canadian journal of plant science 86.3: 647-662.
- Adugna W (2000) Assessment of tissue culture drived regenerants of linseed (Linum usitatissimum L.) in Ethiopia. M.S.C. thesis, Department of Plant Breeding, Faculty of Agriculture, University of the Free State, Bloemfontein, South Africa.
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at Google Scholar Crossref
Citation: Mohammed T (2024) Performance Evaluation of Linseed (Linumusitatissimum L.) Varieties in Buno Bedele Zone, South Western Oromia, Ethiopia.Adv Crop Sci Tech 12: 723.
Copyright: © 2024 Mohammed T. This is an open-access article distributed underthe terms of the Creative Commons Attribution License, which permits unrestricteduse, distribution, and reproduction in any medium, provided the original author andsource are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 1914
- [From(publication date): 0-2024 - Dec 21, 2025]
- Breakdown by view type
- HTML page views: 1605
- PDF downloads: 309