Pathologist-Guided Approach to Deep Learning Prediction of Pediatric Posterior Fossa Tumor Histology
Received: 15-Apr-2022 / Manuscript No. JCEP-22-61133 / Editor assigned: 18-Apr-2022 / PreQC No. JCEP-22-61133 (PQ) / Reviewed: 29-Apr-2022 / QC No. JCEP-22-61133 / Revised: 06-May-2022 / Manuscript No. JCEP-22-61133(R) / Published Date: 13-May-2022 DOI: 10.4172/2161-0681-22.12.411
Abstract
Background: CNS tumors remain among the most frequently discordant pathologic diagnoses in the field of pediatrics. In this study, we examined neuropathologist-guided deep learning strategies towards automation of tumor histology diagnosis targeting the three most common pediatric Posterior Fossa (PF) tumors.
Methods: A retrospective chart review identified 252 pediatric patients with histologically confirmed PF Pilocytic Astrocytoma (PA); Ependymoma (EP); medulloblastoma (MB) across two independent institutions: Site 1: PA(n=87); EP(n=42); MB(n=50); Site 2: PA(n=36); EP(n=9); MB(n=28). The dataset comprised images of tumor-relevant regions captured by neuropathologists while viewing histology slides at 20 × magnification at the microscope. A Resnet-18 architecture was used to develop a 2D deep learning models and to assess model generalization across the two sites. Holdout test set was used to assess each of the model performance.
Results: Model trained exclusively on Site 1 cohort, achieved an accuracy of 0.75 and a F1 score of 0.61 on test set from Site 1; and an accuracy of 0.89 and F1 score of 0.77 on Site 2. Fine tuning on a subset of cohort from Site 2 did not significantly improve model performance.
Conclusion: We demonstrate a potential role implementing AI for histologic diagnosis of the three most common pediatric PF tumors that can generalize across centres. Further, we identify feasibility of AI learning that uses histology images captured by neuropathologists at the microscope and thereby incorporate expert human behavior. Future study could examine AI model developments that use tumor segmentations of histology slides in comparison to expert pathologist-guided image capture as forms of tumor labels.
Keywords: Central Nervous System (CNS); Posterior fossa; medulloblastoma; Pilocytic astrocytoma; Ependymoma
Introduction
Tumors of the Central Nervous System (CNS) collectively represent the most common solid tumors of childhood, accounting for 15%-20% of pediatric malignancies [1]. Despite advances in modern medicine, CNS tumors represent the most common cause of cancer-related deaths in children [1]. Early and accurate tumor diagnosis remains critical to treatment planning, to family counseling, and ultimately, to improving outcomes [2,3]. While imaging offers potential for pre-surgical diagnosis, final tumor diagnosis relies on histopathology of surgical specimens.
Despite the high incidence of CNS tumors among solid childhood cancers, CNS tumors nevertheless are relatively rare, with an incidence rate of 5.7 per 100,000 person-years in the United States [4]. This presents challenges for practicing pathologists outside of major tertiary care referral centers, who may infrequently encounter these tumors.
It is also well-known that discordances in histologic diagnosis of pediatric pathologies are not infrequent upon second review, with CNS tumors among the most frequently discordant pathologic diagnoses in the field of pediatrics [5,6]. Potential contributing factors include local pathologist perception of the cases as being difficult; unusual histological features; discordance between clinical presentation and histology; and incorporation of molecular testing in the updated WHO classification system [6].
Given the recent advances in computer vision applied in medicine, we sought to use deep learning strategies in the automation of tumor histology diagnosis. We targeted Posterior Fossa (PF) tumors, specifically, medulloblastoma (MB), Pilocytic Astrocytoma (PA), and Ependymoma (EP), given their relatively more common occurrence among CNS tumors in children. Further, given the well-known challenges of AI training inherent in histology due to background noise and large histology slide sizes, we proposed the use of screen shot images of relevant tumor regions captured by clinical pathologists at the microscope for AI training.
Materials and Methods
Study cohort
We conducted a retrospective study across two independent institutions (Stanford University [Site 1] and Vanderbilt University [Site 2]) after the IRB approval. The inclusion criteria were
• Pathologically confirmed diagnosis of the following pediatric posterior fossa tumors: medulloblastoma, pilocytic astrocytoma, or ependymoma,
• Patients were aged 1 day to 19 years, and
• H and E glass histology slides were available for review by a neuropathologist. Patients were excluded if the tumor histology diagnosis was unclear.
Histology dataset
Neuropathologists from each site independently viewed individual histology slides under a microscope at 20 × and captured 4800 × 3600 pixel screenshot images with 72 × 72 dpi resolution centred over a tissue region representative of the brain tumor. Effort was made to reduce image capture of normal tissue, white space, and processing artifacts.
Dataset distribution for training
The data were stratified by tumor type to ensure an equal distribution of tumor types in both the training set and validation set. For each site, 80% of the data served as training and 20% was withheld from the training set to serve as a test set to assess the final model performance.
Experimental overview
We conducted the following experimental approaches:
Phase 1: Develop a deep learning algorithm using solely Site 1 data and test its performance on test sets from Site 1 and Site 2.
Phase 2: Fine-tune the best performing model from Phase 1 using a subset of the Site 2 cohort and assess model performance on test sets from Site 1 and Site 2.
Model architecture
We used ResNet architectural backbone pretrained on the Image Net dataset, a compilation of over 14 million images of everyday objects [7,8]. Due to the relatively small cohort size, we used the smallest available pretrained architecture with 18 layers to reduce risk of overfitting. The pretrained ResNet-18 architecture was modified to classify the three PF tumor classes: PA, EP, MB.
Image pre-processing
Pixel values were normalized per PyTorch pretrained model guidelines [9]. All images contained three (i.e., RGB) color original image. In addition to these rescaling options, each image in the train set had a 50% probability of vertical or horizontal flip. Validation and test set images were rescalechannels. We performed several data augmentations for training. Each image used for model training had a 50% probability of rescaling to 224 × 224 dimensions or random cropping of an unmagnified 224 × 224 sizedd to 224 × 224 to allow the model to analyze the image but were not otherwise manipulated; no data augmentations were applied to validation or test set images.
Model training
All models were trained using the Python 3.6 programming language and the PyTorch deep learning framework and a NVIDIA TitanXp Graphic Processing Unit with 12 GB of memory [9]. During training, all layers of the model, including the pretrained convolutional layers, were fine-tuned on the histology training data and trained to minimize classification cross entropy loss. The Adam optimizer was used to update the weights of the model with each iteration [10]. We conducted a two-phase experimental approach, as shown in Figure 1.
Phase 1: Develop a deep learning algorithm using solely Site 1 data and test its performance on test sets from Site 1 and Site 2. In Phase 1, during which the model only had access to Site 1 training data, the model was trained for 10 epochs with a batch size of 64 images and a learning rate of 0.001. Random majority subset (80%) of data from Site 2 served as the test set.
Phase 2: Fine-tune the best performing model from Phase 1 using a subset cohort from Site 2 and assess model performance on test sets from Site 1 and Site 2. In Phase 2, during which the model was further fine-tuned on Site 2 training data, the model was trained for 5 epochs with a batch size of 64 images and a learning rate of 0.0001. Here, a random minority subset (20%) of data from Site 2 was used to finetune the best performing model from Phase 1. Similar to Phase 1, the majority subset (80%) from Site 2 served as test set to determine the model performance.
Each model was trained with 5-fold cross validation, using a 20% proportion of the training set as the validation set. During final evaluation, the model with the lowest total loss was evaluated on the test set to gauge performance.
Statistics
We used a McNemar test to determine model performance between a model trained only on the Site 1 cohort versus a model trained on Site 1 and fine-tuned with the Site 2 cohort.
Results
Study cohort
A total of 252 subjects met the inclusion criteria: 179 from Site 1 and 73 from Site 2. The tumors distributions were as follows. Site 1: PA (n=87); EP (n=42); MB (n=50); Site 2: PA (n=36); EP (n=9); MB (n=28). Total image counts by institution are shown in Table 1 and the data distribution for Phase 1 and 2 experiments are shown in Tables 2 and 3, respectively.
| Stanford | Vanderbilt | Total | |
|---|---|---|---|
| Pilocytic astrocytoma | 87 | 36 | 123 |
| Medulloblastoma | 50 | 28 | 78 |
| Ependymoma | 42 | 9 | 51 |
Table 1: Total image counts by institution.
| Stanford training set | Stanford test set | Vanderbilt test set | |
|---|---|---|---|
| Pilocytic astrocytoma | 69 | 18 | 29 |
| Medulloblastoma | 40 | 10 | 23 |
| Ependymoma | 34 | 8 | 7 |
Table 2: Total image counts in training and test sets for Phase 1.
| Stanford training set | Vanderbilt training set | Stanford test set | Vanderbilt test set | |
|---|---|---|---|---|
| Pilocytic astrocytoma | 69 | 7 | 18 | 29 |
| Medulloblastoma | 40 | 5 | 10 | 23 |
| Ependymoma | 34 | 2 | 8 | 7 |
Table 3: Total image counts in training and test sets for Phase 2.
Model performance
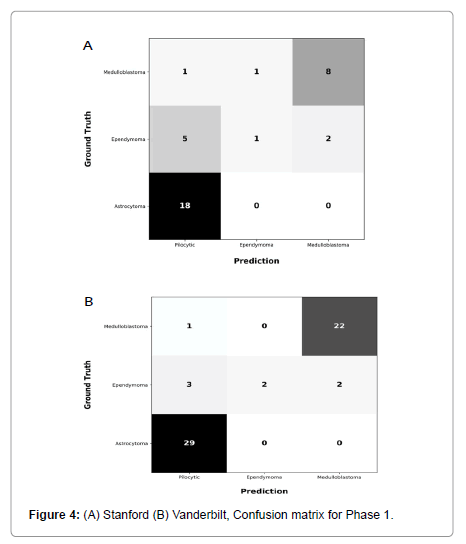
Phase 1: Model trained exclusively on Site 1 cohort, achieved an accuracy of 0.75 and a F1 score of 0.61 on holdout test set from Site 1. This model achieved an accuracy of 0.89 and F1 score of 0.77 on previously unseen cohort from Site 2.
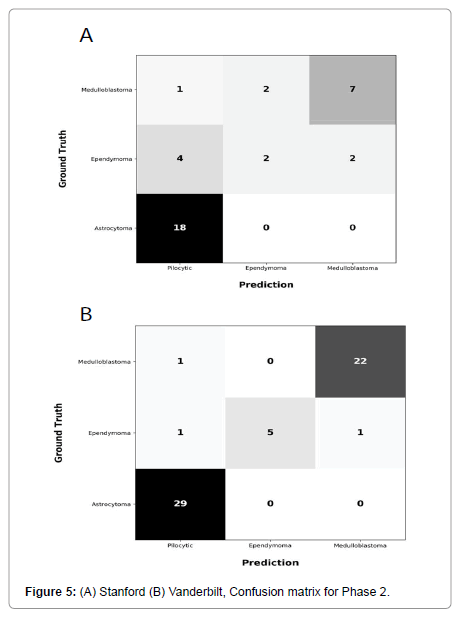
Phase 2: The second model, i.e., model from Phase 1 that is finetuned with a subset of data from Site 2, achieved an accuracy of 0.75 and a F1 score of 0.65 on holdout test set from Site 1. This model achieved an accuracy of 0.95 and F1 score of 0.92 when tested on holdout test set from Site 2.
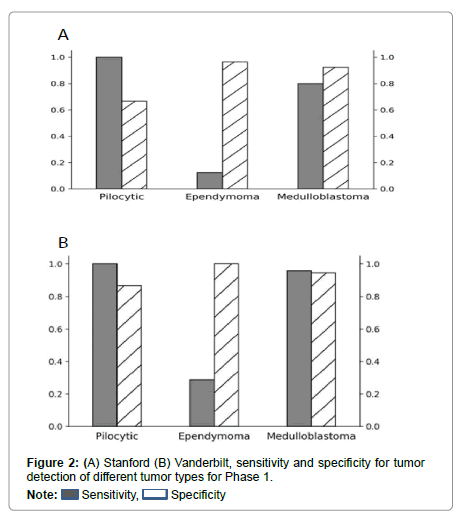
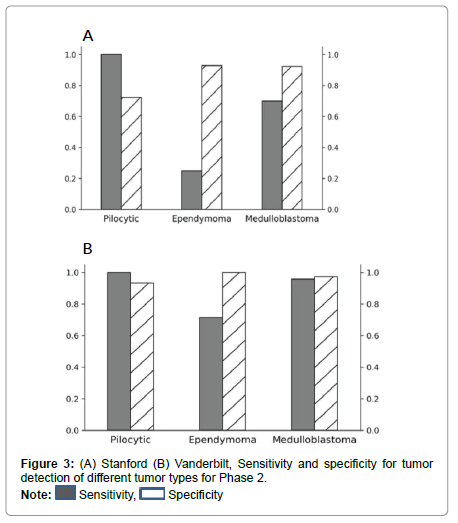
Performance metrics are summarized in Figures 2-5.
Figures 2 and 3 show bar plots comparing sensitivity and specificity for prediction on individual tumor pathologies from Phase 1 and Phase 2 experiments, respectively. Figures 4 and 5 illustrate confusion matrices that showcase model prediction against ground truth labels for each tumor pathology from the test set from Phase 1 and Phase 2 experiments, respectively.
The McNemar test comparing the performance of these two classifiers had a value of 1.0 and a p-value of 1.0 on the test set from Site 1. For the Site 2 test set, the McNemar test was found to have a value of 0.0 and a p-value of 0.25. These findings suggest that the performance of the model fine-tuned with a small subset of external data did not significantly differ from the model trained on data from a single institution.
Results and Discussion
Investigations of CNS tumors have dramatically increased in the past decade with new insights into molecular biology, improvements in imaging, as well as increased utilization of immunohistochemical biomarkers. While various modalities often play complementary roles for diagnosis, histopathology remains fundamental to tumor diagnosis and treatment planning. With a trend toward the digitization of pathology slides, pathology and laboratory medicine as a specialty is uniquely enriched with opportunities for integrating AI-based tools that assist and educate pathologists, enhance workflow efficiency and contribute to precision in diagnosis [11].
Despite potential roles for AI in pathology, large data sizes of high-resolution pathology slides and general lack of annotations pose hurdles to AI model development to a greater extent than in radiology. Prior studies have applied deep learning strategies on histology of breast cancer or lymphoma for more precise evaluation [12,13], but at present, no study has examined the potential role of AI on pediatric brain tumors. Here, we present a histology-based deep learning model predictive of the three most common pediatric PF tumors. Rather than the use of labor-intensive segmentations over tumor boundaries, we used images of diagnostically relevant tumor regions over pathology slides that expert neuropathologist captured while viewing at the microscope and thereby mirrored real world behavior of pathologists. We further assessed generalizability of this approach to model development.
Our base model trained exclusively on Site 1 cohort, achieved an accuracy that ranged 75-89%, with an F1 score that ranged 61%-77% when evaluated on holdout test sets from Sites 1 and 2.
Overall, our pilot results point to a potential AI role for augmenting pathologic diagnosis that can either serve as a “second look” for general pathologists who may be less familiar with rarer tumor histology5,6, or even as an educational tool for pathologists-in-training. We show that this is feasible even when using pathologist-guided images captured at the microscope, without the use of digitized images derived from glass histology slides and without tumor margin segmentations.
Despite promising results, the performance metrics do suggest room for improvement. One contributor might relate to variations in histology slides across institutions, including artifacts and slide preparation differences. Examples of artifacts might include tissue folds, blurred regions, and shadowing, which could confer unpredictable effects on model training [14,15]. Differences in slide preparation, such as slice thickness variation or length of staining time, the so-called “batch effects,” add additional variability [14,15]. Nevertheless, high predictive performance on Site 2 suggests model durability. Since finetuning on external data (Site 2) did not alter model performance, it is possible pixel-based relevant features did not significantly differ across the two sites.
There are several limitations to this study. Our sample size was small, which in part relates to the inherent rarity of pediatric brain tumors. This also explains lower predictive performance of EP tumors, which were fewer compared to PA and MB tumors. To mitigate this problem, we used pathologist-driven, human-guided images of relevant tumor histological regions captured at the microscope, rather than tumor segmentation methods, the latter being typically used in AI training. We also combined two-institution data to assess model generalizability. While the use of raw digitized histology slides could add value, this was not possible given our relatively small cohort size and large amount of noise (e.g., normal tissue, white spaces, and processing artifacts) inherent within the raw histology data.
Conclusion
This Study demonstrates the feasibility of AI learning using images captured by expert neuropathologist at the microscope that mimics the real-world behavior of pathologists as a form of tumor labeling, rather than the use of manual tumor segmentations. Our model demonstrates a level of accuracy, sensitivity and specificity that, while certainly not suited to completely replace expert neuropathologist, could function as an adjunctive tool for general pathologists or students training in pathology seeking a supplemental tool. Future study could examine AI model developments that use tumor segmentations of histology slides in comparison to expert pathologist-guided image capture as forms of tumor labeling.
References
- Udaka YT, Packer RJ (2018) Pediatric brain tumors. Neurol Clin 36: 533-556.
[Cross ref] [Google Scholar] [PubMed]
- Louis DN, Perry A, Reifenberger G, Deimling AV, Figarella-Branger D, et al. (2016) The 2016 World Health Organization classification of tumors of the Central Nervous System: A summary. Acta Neuropathol 131:803-820.
[Cross ref] [Google Scholar] [PubMed]
- Segal D, Karajannis MA (2016) Pediatric brain tumors: An update. Curr Probl Pediatr Adolesc Health Care, 46: 242-250.
[Cross ref] [Google Scholar] [PubMed]
- Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, et al. (2016) CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the united states in 2009-2013. Neuro Oncol 18: v1-v75.
[Cross ref] [Google Scholar] [PubMed]
- Pollack IF, Boyett JM, Yates AJ, Burger PC, Gilles FH, et al. (2003) The influence of central review on outcome associations in childhood malignant gliomas: Results from the CCG-945 experience. Neuro Oncol 5: 197-207.
[Cross ref] [Google Scholar] [PubMed]
- Merabi Z, Boulos F, Santiago T, Jenkins J, Abboud M, et al. (2018) Pediatric cancer pathology review from a single institution: Neuropathology expert opinion is essential for accurate diagnosis of pediatric brain tumors. Pediatr Blood Cancer, 65.
[Cross ref] [Google Scholar] [PubMed]
- He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. IEEE Trans Med Imag 770-778.
- Deng J, Dong W, Socher R, Li L, Kai Li, et al. (2009) ImageNet: A large-scale hierarchical image database. IEEE Trans Med Imag 248-255.
- Paszke A, Gross S, Massa F, Lerer A, Bradbury J, et al. (2019) PyTorch: An Imperative style, high-performance deep learning library. arXiv 1-12.
- Kingma DP, Ba J (2014) Adam: A Method for stochastic optimization. arXiv.
- Serag A, Ion-margineanu A, Qureshi H, McMillan R, Martin MS, et al. (2019) Translational AI and deep learning in diagnostic pathology. Front Med 6: 185.
[Cross ref] [Google Scholar] [PubMed]
- Mi W, Li J, Guo Y, Guo Y, Ren X, Liang Z, et al. (2021) Deep learning-based multi-class classification of breast digital pathology images. CMAR 13: 4605-4617.
[Cross ref] [Google Scholar] [PubMed]
- Achi HE, Belousova T, Chen L, Wahed A, Wang I, et al. (2019) Automated diagnosis of lymphoma with digital pathology images using deep learning. Ann Clin Lab Sci 49: 153-160.
[Google Scholar] [PubMed]
- Toro OJD, Atzori M, Otálora S, Rönnquist P, Müller H, et al. (2019) Convolutional neural networks for an automatic classification of prostate tissue slides with high-grade Gleason score. NPJ Digit Med 19: 113.
[Cross ref] [Google Scholar] [PubMed]
- Kothari S, Phan JH, Stokes TH, Wang MD (2013) Pathology imaging informatics for quantitative analysis of whole-slide images. J Am Med Inform Assoc 20: 1099-1108.
[Cross ref] [Google Scholar] [PubMed]
Citation: Campion AW, Bala WA, Tam L, Lavezo J, Harmsen H, et al. (2022) Pathologist-Guided Approach to Deep Learning Prediction of Pediatric Posterior Fossa Tumor Histology. J Clin Exp Pathol 12: 411. DOI: 10.4172/2161-0681-22.12.411
Copyright: © 2022 Campion AW, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 3692
- [From(publication date): 0-2022 - Apr 02, 2025]
- Breakdown by view type
- HTML page views: 3191
- PDF downloads: 501