Research Article Open Access
Optimization of Phenol Degradation Using Pseudomonas aeruginosa (MTCC 7814) by Plackett-Burman Design and Response Surface Methodology
| Pandimadevi M*, Venkatesh Prabhu M and Vinod Kumar V | |
| Department of Biotechnology, SRM University, India | |
| Corresponding Author : | Pandimadevi M Department of Biotechnology SRM University, India Tel: +91 9444145987 E-mail: pandimadevi.m@ktr.srmuniv.ac.in, pandimadevi2008@gmail.com |
| Received August 13, 2014; Accepted November 25, 2014; Published November 28, 2014 | |
| Citation: Pandimadevi M, Venkatesh Prabhu M, Vinod Kumar V (2014) Optimization of Phenol Degradation Using Pseudomonas aeruginosa (MTCC 7814) by Plackett- Burman Design and Response Surface Methodology. J Bioremed Biodeg 5:261. doi:10.4172/2155-6199.1000261 | |
| Copyright: © 2014 Pandimadevi M, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
|
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
In the present study, statistical screening and optimization of phenol degradation was done by Pseudomonas Aeruginosa (MTCC 7814), which can utilize phenol as a sole carbon and energy source. Nine medium variables Phenol, K2HPO4, K2H2PO4, MgSO4, (NH4)2SO4, MnSO4, FeSO4, NaCl and H3BO3 were screened by Plackett-Burman (PB) method. K2HPO4, (NH4)2SO4, MnSO4 and phenol were significant in PB method. Central composite design (CCD) and Response Surface Methodology (RSM) were applied to optimize the significant variables identified from the PB experiment. Statistical analysis of the experimental results showed optimal values were found to be KH2PO4 0.025 g/L, (NH4)2SO4 0.45 g/L, MnSO4 0.05 g/L and phenol 1 g/L with maximum phenol degradation of 83.86%. Maximum phenol degradation of 81.62% was observed in the validation experiment. This experimental result explained the model was fitted 97.32 % as compare with the result predicted by response surface method. This study indicated the excellent ability of Pseudomonas Aeruginosa to degrade phenol of high concentration.
| Keywords |
| Phenol; Biodegradation; Pseudomonas aeruginosa; Plackett-Burman Design; Central composite design |
| Introduction |
| Phenol is one of the most common toxic environmental pollutants, arising during the processing of resins, plastics, dyes, varnishes, pharmaceuticals and pesticides from several industrial processes and several chemical industries [1]. Phenol concentrations are toxic to fish if it is higher than 2 mg/L and concentrations between 10 and 100 mg/L would result in death of many aquatic lives within 4 days [2]. The Environmental Protection Agency (EPA) stated that less than 1.0 ppb of phenols in surface waters is good for health. The World Health Organization (WHO) has set a limit level of 1 mg/L to regulate the phenol concentration in drinking waters [3]. |
| Therefore, development of new methods has generated significant interest for phenol removal from industrial wastewater [4]. Chemical or physical methods for phenolic waste treatment were used conventionally such as chemical oxidation, solvent extraction and adsorption, but these processes are facing secondary effluent problems. Apart of these methods, biological treatment i.e. Biodegradation is good alternative. Biodegradation is versatile, inexpensive and can potentially turn a toxic material into harmless products [5]. |
| The use of pure cultures microorganisms, especially for efficient metabolism of the contaminant, is advisable as a better alternative compare to mixed culture [6-9]. In general, the one-variable at a time (OVAT) approach in optimization studies is not difficult, but it decreases accuracy of the effects of interacting factors and might lead to misinterpretations of the results. On the other hand, statistical planned experiments are better to minimize the error, to determine the effect of parameters and to achieve results in an economical manner [10]. Statistical experimental designs such as Plackett-Burman and response surface methodology (RSM) [11] can optimize all the significant parameters effectively [10]. Plackett-Burman design provides a fast and effective way to identify the important factors among a large number of variables by saving time and maintaining important information on each parameter. Response Surface Methodology, which is supported by software, is an empirical technique derives for the evaluation of the relationship of a controlled experimental set of factors and observed results. Basically, this optimization process involves collection of mathematical tools to create, develop and refine the model [5,12,13]. The aim of the present work was to screen and optimize the medium components by statistical methods for phenol degradation by Pseudomonas Aeruginosa MTCC 7814. |
| Materials and Methods |
| Bacterial Strain |
| The bacterium P. Aeruginosa (MTCC 7814) was ordered from Microbial Type Culture Collection and Gene Bank, Institute of Microbiology (IMTECH), Chandigarh, India, in lyophilized form. Stock cultures were then obtained by standard spread plate microbial techniques. The microorganisms were maintained on nutrient agar slant and stored at 300C for further use. Inoculum was prepared by transferring two lapful’s of the microorganism from nutrient agar slant into a 250-mL Erlenmeyer flask containing 100 mL LB liquid medium. The flask was incubated in a shaking at 34°C and 120 rpm for 24 hr. |
| Medium |
| Phenol (99% purity) was purchased from M/s HIMEDIA Pvt. Ltd., India. 4-amino antipyrine and potassium ferricyanide were purchased from M/s RANKEM Pvt. Ltd., India. To avoid photo-oxidation, 10% stock solution of phenol was stored in brown glass bottles. The growth medium contained phenol as the sole carbon source [14]. The revived cultures were first grown in mineral salt medium was modified from the one suggested by [15]. The medium was sterilized in two parts to avoid precipitation of ferrous salts during autoclaving. These were denoted as Part A and Part B with the following compositions: |
| Part A K2HPO4,, KH2PO4, FeSO4.7H2O |
| Part B (NH4)2SO4, MgSO4·7H2O, MnSO4, H3BO3, NaCl |
| After autoclaving, the contents were cooled and then mixed together and kept as stock solution from which known quantities were taken for the cultivation of the microorganisms. These two parts and phenol were mixed before start of any batch experiment. The phenol solution was prepared with the concentration ranging from 200 mg/l to 500 mg/l. The pH of the medium thus obtained was 7.0±0.1. |
| Acclimatization |
| The primary culture was prepared by transferring the two loops full of microorganisms from an agar slant culture into 100 ml of feed medium containing 25 ml of mineral salt and 75 ml of 50 mg phenol in a series of 250 ml sterile conical flasks and the period of incubation was repeated. This was the secondary culture that was used as the inoculums for the degradation studies as this ensures that the organisms had fully adapted to growth on the phenol, as a sole source of carbon energy and all the inoculum transfers were done in exponential phase [16]. The temperature in all the batch experiments was maintained at 34â°C. For each concentration, three flasks of 250 ml capacity with 100 ml working volume were kept in incubator-cum shaker. At the most, 20 ml were taken from each flask. All the transfers were made in UV chamber, and glass wares and medium properly autoclaved. The batch experiments were repeated and the results were found reproducible within acceptable range. |
| Phenol Estimation |
| The concentration of phenol undegraded in the solution was determined by a UV-vis spectrophotometer using 4-amino antipyrine as a color reagent [17,18]. For measuring biomass, the samples were centrifuged at approximately 6000 rpm for 20 min. The supernatant was used for phenol determination. The biomass was resuspended in distilled water and optical density of this suspension was measured against distilled water as reference at 600 nm using UV-Visible double beam spectrophotometer. All experiments were performed in triplicates and the average of the three independent experiments was taken as the result. |
| Plackett-Burman design |
| Plackett-Burman design, an efficient way to identify the important factors among a large number of variables [19], was used in the present study to screen the important variables that significantly influenced phenol degradation. In this study, a 12-run Plackett-Burman design was applied to evaluate nine factors (including two dummy variables). K2HPO4, KH2PO4, MgSO4, (NH4)2SO4, MnSO4, FeSO4, H3BO3, NaCl and phenol were selected for the screening process by PB design. Each variable was examined at two levels: (-1) for the low level and (+1) for the high level. Each row represents different experiment and each column represents different variables (Table 1). |
| The effect of individual variable on phenol degradation was calculated by the following Equation: |
| Σ (Xi) = 2(ΣMi+ -Mi-)/N (1) |
| Where, Σ (Xi) is the effect of the tested variable (Xi), N is the no. of trials and Mi+ and Mi- are responses (phenol degradation) of trials at which the variable is at its high or low levels respectively [20]. |
| Response Surface Methodology |
| The optimal levels of the significant factors and the interactions of these variables on phenol degradation were analyzed by CCD [21]. In this study, a four-factor; five-level CCD with 31 runs was employed. Tested variables (KH2PO4 concentration, (NH4)2SO4 concentration, MnSO4 concentration, Phenol concentration) were denoted as X1, X2, X3, and X4, respectively, and each of them was kept at five different levels, combining factorial points (–1, +1), axial points (–2, +2), and central point (0), as shown in Table 3. In this design, cube points were 16, center points in cube were 7, axial points were 8 and alpha level 2 was kept in consideration. |
| The second-order model used to fit the response to the independent variables is shown below: |
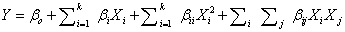 (2) (2) |
| Where, Y is the predicted response (phenol degradation); Xi and Xj are input variables that influence the response Y; k is the number of variables; β0 is the constant term; βi is the ith linear coefficient; βii is the ith quadratic coefficient; βij is the ijth interaction coefficient. |
| Analysis of variance (ANOVA) was conducted to determine the significance of model and regression coefficients and it was shown in the Table 6. The quality of polynomial equation was judged by determination coefficient (R2), and its statistical significance was checked by Fischer’s F-test. The significance of regression coefficients was tested by Student’s t-test. The response contour plots of the modelpredicted responses were utilized to assess the interactive relationships between the significant variables. |
| Data analysis |
| MINITAB© Version 14 was used for designing experiments as well as for regression and graphical analysis of the experimental data obtained. |
| Results and Discussion |
| Screening of important variables using PB design |
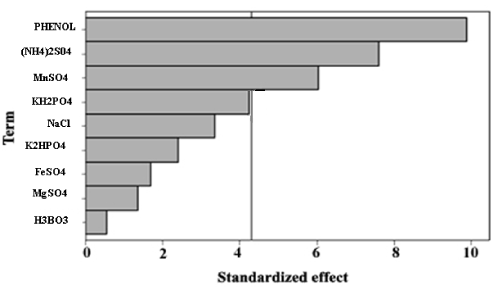
| The data listed in Table 2 indicated a wide variation in phenol degradation, from 51.43% to 75.34%, in the 12 trials. The variation suggested that process optimization was important for improving the removal efficiency of phenol. Analysis of the regression coefficients and the t-values of 9 factors (Table 5) showed that C, E, F and J had positive effects on phenol degradation, whereas A, B, D, G, H had negative effects. The variable with confidence level above 95% is considered as significant parameter. It was clear that variables C, E, F and J were the significant factors, while variables A, B, D, G and H, with confidence levels below 95%, were considered insignificant and were not included in CCD experiments. The Pareto chart shows the significant variables in Figure 1. |
| The model equation for phenol degradation (Y) could be written as: |
| Y = 82.53 – 1.14 A – 2.43 B -11.53 C + 0.56 D + 25.75 E + 61.27 F – 13.72 G – 6.80 H – 0.33 J |
| Optimization by response surface methodology and regression analysis |
| CCD was employed to study the interactions between the significant factors and also to determine their optimal levels. The design matrix of tested variables and the experimental results are represented in Tables 4 and 5. Multiple regression analysis was used to analyze the data and thus a second-order polynomial equation was derived, as follows: |
| Y = 67.48 – 4.49X1 – 1.55 X2 -1.64X3 – 7.09X4 +1.00 X1 2 + 1.04 X2 2– 0.59 X3 2 -0.11X4 2 – 1.64 X1X2 + 1.96 X1X3 – 0.44 X1X4 + 0.93 X2X3 + 0.44 X2X4 - 0.44 X2X4 |
| From the Table 6, the ANOVA for the CCD, it was found that the pred-R2 of 0.877 was in reasonable agreement with adj-R2 of 0.76. The adequate precision, the signal to noise ratio of 30.303, suggested an adequate signal. The “Lack of Fit F-value” of 1.45 implied that the Lack of Fit was not significant relative to the pure error. There was a 33.7% chance that a large “Lack of Fit F-value” could occur due to noise. |
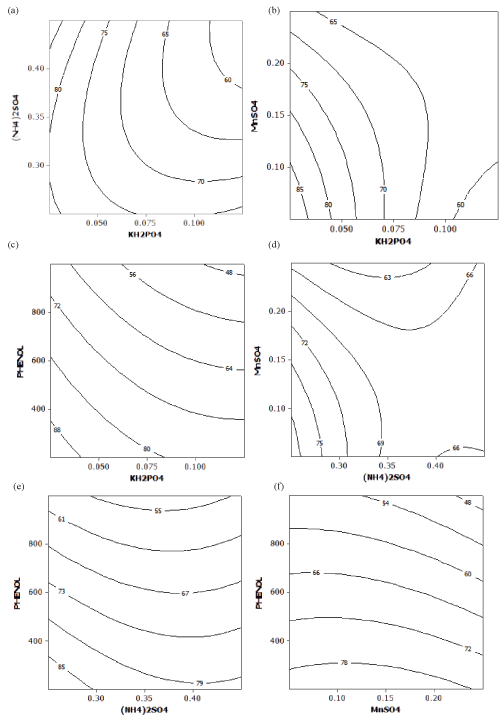
| Contour Plots |
| The contour plots help to visualize the shape of a response surface. When the contour plot displays ellipses or circles, the center of the system refers to a point of maximum or minimum response [21]. The interaction effect between pair of variables on phenol degradation was well understood by the contour plots represented in Figure 2a–f by keeping the other two factors constant at its middle level. The hold values of KH2PO4, (NH4)2SO4, MnSO4 and phenol were 0.075, 0.35, 0.15 and 0.6 g/L respectively. Figure 2a-f showed maximum phenol degradation was observed at the middle level of each pair of variables. |
| Optimization of Phenol Degradation |
| The response optimizer tool was used to predict the optimal medium concentration on phenol degradation. The optimal values were found to be KH2PO4 0.05 g/L, (NH4)2SO4 0.35 g/L, MnSO4 0.15 g/L and phenol 500 mg/L with predicted phenol concentration 83.86 %. The composite desirability of 67.6 % shows the accuracy of the model. |
| Validation of the Model |
| In order to validate the model, all four significant variables were kept on optimized concentrations. The experiment was conducted on triplicates. The experimental phenol degradation was found to be 81.62 % which shows that the model was fitted with 97.32% accuracy. It means the model was significant. |
| Conclusion |
| This works attempts for the higher percentage degradation of high concentration phenol. Plackett Burman method was employed to identify the significant variables that influence the degradation of phenol. Four medium variables out of nine were identified to be significant on phenol degradation by PB experiment. The results obtained from RSM were clearly explained that optimum values of significant variables had a significant effect and promotes an increase in percentage of phenol degradation. The optimization results shows that the maximum phenol degradation of 81.62 (%) was obtained when KH2PO4, (NH4)2SO4, MnSO4 and phenol were kept on optimized concentrations 0.05 g/L, 0.35 g/L, 0.15 g/L and 500 mg/L respectively. Result of the validation experiment under optimal conditions was 97.32 % fitted to the values predicted by the software. |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 |
| Table 4 | Table 5 | Table 6 |
Figures at a glance
 |
 |
| Figure 1 | Figure 2 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 13775
- [From(publication date):
December-2014 - Nov 21, 2024] - Breakdown by view type
- HTML page views : 9343
- PDF downloads : 4432
