Research Article Open Access
Occurrence of Cellulose Degraders in Fruit and Vegetable Decaying Wastes
Jaya Philip1*, Tanuja T2 and Bedi S3
1Department of Industrial Microbiology, Patna Women’s College, Patna University, Patna, Bihar, India
2Department of Botany, TPS College, Magadh University, Bodhgaya, Bihar, India
3Department of Botany, Patna Women’s College, Patna University, Patna, Bihar, India
- *Corresponding Author:
- Jaya Philip
Department of Industrial Microbiology
Patna Women’s College, Patna University
Patna, Bihar, India
Tel: 7544852759
E-mail: jayaphilipmicrobio@gmail.com
Received Date: October 20, 2016; Accepted Date: October 28, 2016; Published Date: October 31, 2016
Citation: Philip J, Tanuja T, Bedi S (2016) Occurrence of Cellulose Degraders in Fruit and Vegetable Decaying Wastes. J Bioremediat Biodegrad 7:373. doi: 10.4172/2155-6199.1000373
Copyright: © 2016 Philip J, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
The present study aims at isolation and characterisation of cellulose degrading bacterial and fungal strains from fruit and vegetable decaying waste such a banana peel, sugarcane bagasse, coconut coir, citrus wastes, potato peel, sponge guard, cucumber peel. 23 bacterial colonies and 12 fungal colonies were obtained on Nutrient Agar and PDA respectively by plating the aliquots of fivefold serially diluted decaying samples. 7 bacterial strains and 4 fungal strains were cellulose degraders as screened on CMC agar flooded with iodine. Also, the Z: C ratio of the fungal and bacterial isolates were recorded to see the degradation potential. Further, characterization of cellulose degraders on the basis of culture and morphology was done and for bacterial isolates biochemical tests were also performed. Results of the Biochemical tests were fed into the ABIS online bacterial identification tool and the bacterial isolates were identified as Bacillus subtilis ~98% (acc: 30%), Bacillus licheniformis ~99% (acc: 32%), Streptococcus ~97% (acc:20%), Bacillus smithii 99% (acc: 30%), Bacillus firmus 99% (acc: 33%), Brevibacillus laterosporus 98% (acc: 32%), Pseudomonas chlororaphis 75% (acc: 33%). However, the 16s rRNA sequencing has to be performed for confirmation of the bacterial isolates. While the fungal isolates obtained were identified as Aspergillus niger, Penicillium, Aspergillus flavus and Rhizopus on the basis of cultural and morphological characterization. The isolated bacterial strains will not only help in the production of useful end products from bio-degradation of the abundant, inexpensive and renewable cellulose wastes but also help in the proper disposal of agricultural and domestic wastes.
Keywords
Aspergillus niger; Cellulose; Bioconservation; Biomass
Introduction
Cellulose is the major constituent of plant biomass. It is the most abundant carbohydrate in nature [1] and regarded as an important renewable resource for bioconservation. It is the most common organic polymer, representing about 1.5 × 1012 tons of the total annual biomass production through photosynthesis especially in the tropics and is considered to be an almost inexhaustible source of raw material for different products [2]. Enormous amounts of agricultural, industrial and municipal cellulosic wastes have been accumulating [3]. It has become a topic of considerable economic interests to develop processes for effective treatment and utilization of cellulosic wastes as inexpensive carbon sources. Cellulose provides a key opportunity for achieving tremendous benefits of biomass utilization. Cellulose is commonly degraded by an enzyme called cellulase. The cellulases have great potential in saccharification of lignocellulosics to fermentable sugars which can used for production of bioethanol, lactic acid, and Single Cell Protein (SCP) [4]. Microbial degradation of cellulosic waste and the downstream products resulting from it is accomplished by a concerted action of several enzymes, the most prominent of which are the cellulases, which are produced by a number of microbes and comprise different enzyme classification. Cellulases are bioactive compound which hydrolyze cellulose beta (1,4-D) - glucan linkages and produce as primary products glucose, cellobiose and cello-oligosaccharides [5]. Fungi are the main cellulase producing microbes though a few bacteria have been also reported to yield cellulase activity. Commercial enzymes are expensive, because they are produced from refined substrates and usually patented organisms. It is therefore imperative that cheaper substrates from local sources for enzyme production be investigated and that bacteria with good enzyme producing capacity be locally isolated [6]. The present study was, therefore, an attempt to isolate and characterize cellulose degrading bacterial and fungal strains from fruit and vegetable decaying wastes which can be used for cellulase production.
Materials and Methods
Collection of samples
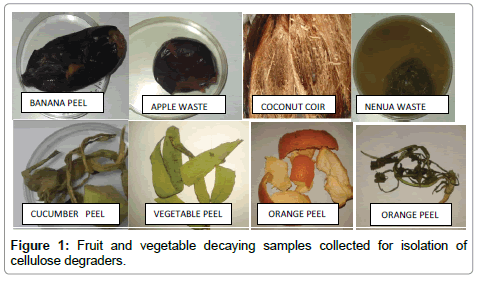
Fruit and vegetable decaying waste samples such as banana peel, sugarcane bagasse, coconut coir, citrus wastes, potato peel, sponge guard and cucumber peel were collected from the kitchen waste and juice outlets (Figure 1).
Media used
For isolation, screening and maintenance of cellulose degrading bacterial strains: Nutrient agar (Peptone, 5 g; Beef extract, 3 g; Sodium Chloride, 5 g; Agar, 15 g; Distilled water, 1000 ml; pH 7.0); Carboxy Methyl cellulose (CMC) agar (CMC, 10 g; Dipotassium hydrogen phosphate, 1 g; Potassium dihydrogen phosphate, 1 g; Magnesium sulphate, 0.2 g; Ammonium nitrate, 1 g; Ferric chloride, 0.05 g; Calcium chloride, 0.02 g; Agar, 20 g; Distilled water, 1000 ml; pH 7.0); Cellulose Congo-red agar media (Dipotassium hydrogen phosphate, 0.5 g; Magnesium sulphate, 0.25 g; Cellulose, 2 g; Congo red, 0.2 g; Gelatin, 2 g; Agar, 15 g; Distilled water, 1000 ml; pH 6.8).
For biochemical and physiological characterization: Fermentation broth (Peptone, 5 g; Beef extract, 3 g; Lactose, 5 g; Glucose, 5 g; Sucrose, 5 g; Sodium chloride, 15 g; Phenol red, 0.018 g; Distilled water, 1000 ml); Simmon’s citrate agar (Ammonium dihydrogen phosphate, 1 g; Dipotassium hydrogen phosphate, 1 g; Sodium chloride, 5 g; Sodium citrate, 2 g; Magnesium sulphate, 0.2 g; Bromothymol blue, 0.08 g; Agar, 15 g; Distilled water, 1000 ml); Tryptone broth (Tryptone, 10 g; Sodium chloride, 55 g; Calcium chloride, 1 ml; Distilled water, 1000 ml); MR-VP broth (Peptone, 7 g; Potassium phosphate, 5 g; Dextrose, 5 g; Distilled water, 1000 ml); SIM agar (Peptone, 30 g; Beef extract, 3 g; Ferrous ammonium sulphate, 0.2 g; Sodium thiosulphate, 0.025 g; Agar, 3 g; Distilled water, 1000 ml).
Chemicals used
For isolation and screening of cellulose degrading bacterial strains: Gram’s iodine solution, Iodine, 1 g; Potassium iodide, 2 g; Distilled water, 300 ml).
For biochemical and physiological characterization: MR indicator (Methyl red, 0.04 g; Ethyl alcohol, 40 ml; Distilled water, 60 ml); VP reagent I (Napthol, 5 g; Ethanol, 95 ml); VP reagent II (Potassium hydroxide, 40 g; Distilled water, 100 ml); Kovac’s reagent (p-Dimethylaminobenzaldehyde, 5 g; Amyl alcohol, 75 ml; Conc. Hydrochloric acid, 25 ml), Catalase reagent (Hydrogen peroxide, 3 g, Distilled water, 100 ml).
Isolation and purification of the bacterial and fungal strains: The samples were serially diluted in sterilized normal saline and the aliquots of fivefold of the soil solutions were plated on the sterilized solidified Nutrient Agar medium and Potato Dextrose Agar medium in the petri plates in aseptic condition. The Nutrient Agar plates were incubated at 37°C for 24 hours for the growth of bacteria and the Potato Dextrose Agar plates were incubated 26°C for 2 to 3 days. The different colonies of bacteria and fungi thus obtained were purified by single streak method and screened for their cellulolytic activities.
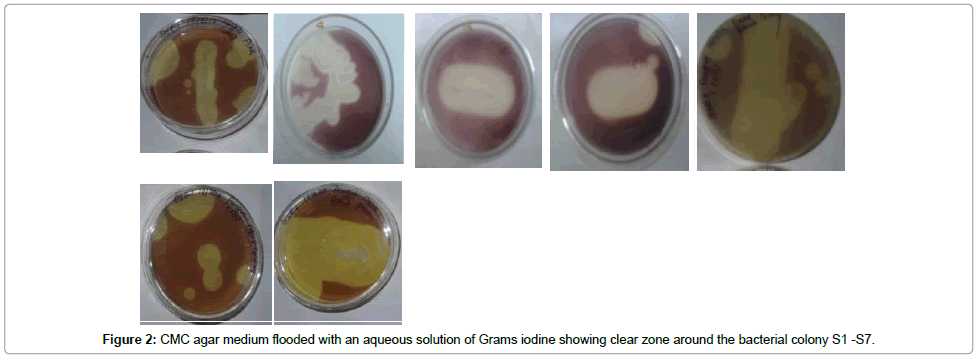
Screening of cellulose degrading microorganisms: The pure fungal cultures were allowed to grow on CMC Agar plates at 26°C for 5 days. CMC Plates streaked with pure bacterial colonies were incubated at 37°C for 5 days to allow the secretion of cellulase and degradation of cellulose present in media in the form of CMC. After incubation CMC agar medium was flooded with an aqueous solution of Grams iodine for 10 minute to visualise the hydrolysis zone. The Grams iodine solution was then poured off. The clear zone was observed around the colonies. The strains showing a clear zone due to utilisation of CMC were selected as potential cellulolytic strains for further study.
Characterization of selected isolates on the basis of cultural, morphology and biochemical tests: The selected bacterial and fungal strains were culturally characterized by observing the colour, texture and margin of the colonies on Nutrient Agar medium and Potato Dextrose Agar medium. Morphological characterization was done by Gram’s staining in case of bacterial strains and lacto-phenol staining for the fungal strains. Then bacterial slides were observed under 100X and the fungal slides were observed under 40X magnification of the research microscope. Further the selected bacterial isolates were subjected to biochemical tests as per Bergey’s Manual of Systematic Bacteriology [7] like carbohydrate fermentation, catalase production, indole production, citrate utilization, MR-VP reaction, hydrogen sulphide production, Growth in 7% NaCl, etc.
Identification of bacterial and fungal isolates: Results of the Biochemical test were fed into the ABIS online bacterial identification tool for identification [8]. While the fungal isolates obtained were identified on the basis of cultural and morphological characterization.
Results and Discussion
Isolation and purification of the microbial strains from the collected soil samples
23 bacterial colonies were obtained on Nutrient Agar and 12 fungal colonies plates by plating the aliquots of fivefold serially diluted decaying samples. The colonies were purified by single streak method on Nutrient Agar and Potato Dextrose Agar plates.
Screening of cellulose degrading microorganisms
Out of 23 bacterial strains isolated 7 were cellulose degraders and out of 12 fungal strains 4 were cellulose degraders. After screening the isolates for cellulose degradation on CMC agar 7 strains marked as S1, S2, S3, S4, S5, S6, and S7 were selected on the basis of area of clear zone around the colonies after flooding with Grams iodine (Figure 2). The ratio of width of the culture growth along the streaked line and width of the zone was calculated to obtain the zonal ratio (Table 1) to study the extent of the enzyme production. More the value of Z as compared to C, more is the enzyme production by that particular bacterial strain. Therefore, according to the observation of the zonal ratio of the bacterial strains on CMC media, S7 shows the maximum Cellulase production. Also, the Z:C ratio of the fungal isolates were recorded as shown in Table 2. Except F1 all other fungal isolates showed similar enzyme activity.
| Strain Number | Width of Zone in cm (Z) | Width of Culture in cm (C) | (Z:C ratio) |
|---|---|---|---|
| S1 | 0.6 | 0.3 | 2:1 |
| S2 | 1.0 | 0.5 | 2:1 |
| S3 | 0.6 | 0.4 | 3:2 |
| S4 | 1.7 | 0.8 | ~2:1 |
| S5 | 0.6 | 0.4 | 3:2 |
| S6 | 0.6 | 0.4 | 3:2 |
| S7 | 0.9 | 0.3 | 3:1 |
Table 1: Zonal ratios of bacterial strain S1- S7 on CMC medium.
| Fungal Strain Number | Width of Zone in cm(Z) | Width of Culture in cm (C) | (Z:C ratio) |
|---|---|---|---|
| F1 | 2.4 | 1.7 | 1:0.7 |
| F2 | 1.2 | 0.7 | 2:1 |
| F3 | 2.6 | 1.3 | 2:1 |
| F4 | 1.6 | 0.8 | 2:1 |
Table 2: Z: C ratio of the fungal isolates F1-F4.
Characterization of selected isolates on the basis of cultural, morphology and biochemical tests
The selected bacterial colonies isolated on NA media (Figure 3) and the fungal colonies isolated on PDA (Figures 4 and 5) from the samples showed the following characteristics as shown in Tables 3 and 4 respectively.
| Bacterial Isolates | Characteristics | ||
|---|---|---|---|
| Colour | Elevation | Texture | |
| S1 | White | Flat | Slimy |
| S2 | Pale yellow | Flat | Slimy |
| S3 | White | Flat | Slimy |
| S4 | Creamish | Convex | Mucoid |
| S5 | Creamish | Convex | Mucoid |
| S6 | Creamish | Flat | Mucoid |
| S7 | Creamish | Flat | Mucoid |
Table 3: Cultural characteristics of bacteria S1-S7 on nutrient agar medium.
| Isolates | Characteristics | |
|---|---|---|
| Colour | Texture | |
| F1 | Black | Powdery |
| F2 | Reddish pink | Cottony |
| F3 | Dark green | Powdery |
| F4 | Black | Cottony |
Table 4: Cultural characteristics of Fungal isolates F1-F4 on Potato Dextrose Agar.
Morphological characteristics of the selected isolates
The selected cellulose degrader was critically examined for its morphology. The Gram’s staining of the bacterial (Figure 5) and cotton blue staining of the fungal isolates (Figure 6) were done and then observed under compound microscope. The morphological characters of the selected bacterial isolates and fungal isolates has been tabulated in Tables 5 and 6 respectively.
| Isolates | Characteristics | |
|---|---|---|
| Gram’s Reaction | Shape | |
| 1 | Positive | Coccusin Chains |
| 2 | Positive | Bacilli(Short Rods) |
| 3 | Positive | Bacilli (ShortRods) |
| 4 | Positive | Bacilli (in Chains) |
| 5 | Positive | Bacilli(Short Rods) |
| 6 | Positive | BacillusSpore Formers |
| 7 | Positive | Bacilliin Chains |
Table 5: Morphological characteristics of Bacterial Strain S1-S7.
| Strain | Characteristics |
|---|---|
| 1 | Single- celled spores (conidia) in chains developing at the end of sterigma arising from the terminal bulb of the conidiophores, the vesicle; long conidiophores arise from a septate mycelium |
| 2 | Single- celled spores (conidia) in chains developing at the end of sterigma arising from the terminal bulb of the conidiophores, the vesicle; long conidiophores arise from a septate mycelium |
| 3 | Single- celled spores (conidia) in chains developed at the end of sterigma arising from the metula of the conidiophores; branching conidiophores arise from a septate mycelium |
| 4 | Single cell spores in chains developing of the end of sterigma arising from the medulla of the conidiophores, branching conidiophores arising from a septate mycelium. |
Table 6: Morphological characteristics of Fungal isolates F1-F4 on Potato Dextrose Agar.
Biochemical characterization of selected bacterial isolates
Strains 1-7 were characterized biochemically and the results were recorded in Table 7.
| Biochemical tests | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| Glucose fermentation | + | + | + | + | + | + | + |
| Sucrose fermentation | - | - | - | - | - | - | + |
| Lactose fermentation | - | - | - | - | - | - | - |
| MR-VP | - | - | - | - | - | - | - |
| Indole Test | - | - | - | - | - | - | - |
| Citrate Utilization | - | - | - | - | - | - | - |
| Catalase Test | + | + | + | + | + | + | + |
| H2S Production | + | - | - | - | - | - | - |
| Growth at 7% NaCl | - | + | - | - | - | - | - |
| Casein hydrolysis | + | + | + | + | + | + | + |
| Starch hydrolysis | + | + | + | + | + | + | + |
| Nitrate reductase | + | + | + | + | + | + | + |
Table 7: Results of biochemical tests of bacterial strains (S1- S7).
Identification of bacterial and fungal isolates
The results of the biochemical test were fed into the ABIS online bacterial identification tool. According to the identification software which the bacterial isolates were S1 as Bacillus subtilis ~98% (acc: 30%), S2 as Bacillus licheniformis ~ 99% (acc: 32%), S3 Streptococcus ~97 (acc: 20%), S4 as Bacillus smithii (99%), S5 as Bacillus firmus (99%), S6 as Brevibacilus laterosporus (98%) and S7 as Pseudomonas chlororaphis (75%), however, the 16s rRNA sequencing has to be performed for confirmation of the bacterial isolates [9,10]. The fungal isolates were Aspergillus niger, Penicillium sps., Aspergillus flavus, Rhizopus sps.
Conclusion
Bacteria are well known agents of decomposition of organic matter in general and of cellulosic substrates in particular [9]. As bacteria, can utilize wide range of cellulosic wastes, therefore, interest in the search for cellulase producing novel bacterial species is increasing. Such habitats which are rich in cellulosic substrates are the best sources in which cellulolytic organisms can be isolated. Further, the wide availability and cost effective sources and an added advantage of ease of isolation as well as processing plays an important role in their selection. The isolated strains will not only help in the production of useful end products from the bio-degradation of the abundant, inexpensive and renewable cellulose but also help in the proper of agricultural, municipal and domestic wastes. In nutshell, the cellulose enzymes will be commonly used in many industrial applications and the demands for more stable, highly active and specific enzymes will be the most stirring technology of future.
References
- Koo YM (2001) Pilot-scale production of cellulase using Trichoderma reesei Rut C-30 in fed-batch mode. J MicrobiolBiotechnol 11: 229-233.
- Klemn D, Schmauder HP, Heinze T (2002) Biopolymers. Volume 6. Vandamme E, Baets D, Steinbuchell A (eds.), pp: 290-292.
- Oberoi HS, Chavan Y, Bansal S, Dhillon GS (2010) Production of cellulases through solid state fermentation using kinnow pulp as a major substrate. Food Bioprocess Technol 3: 528-536.
- Maki M, Leung KT, Qin W (2009) The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int J Biol Sci 5: 500-516.
- Schülein M (1998) Kinetics of fungal cellulases. BiochemSoc Transact 26: 164-167.
- Adeleke EO, Omafuvbe BO, Adewale IO, Bakare MK (2012) Purification and characterisation of a cellulase obtained from cocoa (Theobroma cacao) pod-degrading Bacillus coagulans Co4. Turk J Biochem 37: 222-230.
- Buchanan RE, Gibbons NE (1974) Bergey's Manual of Determinative Bacteriology. Williams & Wilkins Co., Philadelphia, PA, USA.
- Gomashe AV, Gulhane PA, Bezalwar PM (2013) Isolation and screening of cellulose degrading microbes from nagpur region soil. Int J Life Sci 1: 291-293.
- Lynd LR, Weimer PJ, Van Zyl WH, Pretorius IS (2002) Microbial cellulose utilization: fundamentals and biotechnology. MicrobiolMolBiolRev 66: 506-577.
- Bergey D (1957) Manual of Determinative Bacteriology. American Society for Microbiology.7thedn. Williams &Wilkins’s Co. Publishers, Baltimore, USA.
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 14403
- [From(publication date):
November-2016 - Jul 04, 2025] - Breakdown by view type
- HTML page views : 13306
- PDF downloads : 1097