Novel RNA Polymerase I Inhibitor CX-5461 Exhibits Antitumor Activity in Multiple Myeloma
Received: 14-Apr-2022 / Manuscript No. AOT-22-60862 / Editor assigned: 18-Apr-2022 / PreQC No. AOT-22-60862(PQ) / Reviewed: 02-May-2022 / QC No. AOT-22-60862 / Revised: 13-Jun-2022 / Manuscript No. AOT-22-60862(R) / Published Date: 20-Jun-2022
Abstract
rDNA transcription is steadily dysregulated in multiple myeloma, through oncogenes and anti-tumor routes, and particularly by c-Myc. The main downstream goals of c-Myc involve ribosomal biogenesis to enhance the protein translation capacity necessary to support the growth and self-renewal programs of malignancy cells. In the research of therapeutics to improve cancer treatment, the last 10 years have shown a renewed interest in targeting ribosome biogenesis. In the present study, we have demonstrated promise for CX-5461 as a new therapeutic target in multiple myeloma. We report that CX-5461 irreversibly inhibits ribosomal RNA (rRNA) transcription by arresting RNA polymerase I (RPI/Pol1/PolR1) in a transcription initiation complex causing down regulation of 47S and inducing the ribosomal stress. We showed that CX-5461 upregulated p53 pathway, and down regulated c-Myc causing cell cycle arrest and cell death
Keywords: CX-5461; TP53; Multiple myeloma; Cell cycle; 47S
Introduction
The deregulation of ribosome synthesis can be the cause of alterations in the cell cycle and cell growth, leading to cell growth, leading to uncontrolled proliferation, which could uncontrolled proliferation leading to the development of cancers [1]. The deregulation of ribosome synthesis also causes a phenomenon called nucleolar stress which leads most often to a modification of the size and number of and number of nucleoli [2].
Myc (proto-oncogene) overexpression induces the proliferation of myeloma cells by stimulating ribosome synthesis by acting directly on the transcription of pre-rRNAs (via Pol I and Pol III) and the expression of assembly factors and ribosomal proteins [3]. Multiple myeloma is a neoplastic plasma cell disorder; this disease is characterized by a very high overall level of protein synthesis due to the production of monoclonal immunoglobulin [4]. Nevertheless, despite improvements in survival, MM remains incurable and resistance to standard treatments is inevitable that the majority of patients will encounter during their illness [5]. The introduction of various new therapeutic, such as rRNA inhibitor [6].
The links between ribosome production and cell growth make ribosome biogenesis a new strategic target for the treatment of cancer [7]. Inhibition of ribosome synthesis disrupts cell proliferation at two levels directly affecting the cell's ability to synthesize proteins, and indirectly stabilizing the p53 tumor suppressor [8].
In the current study, we aimed to identify the therapeutic potential of RNA pol I inhibition with CX-5461 in multiple myeloma, which is currently in clinical trials. Our studies show that the inhibition of rRNA induces the p53 pathway and decreases c-Myc level expression.
Materials and Methods
Multiple myeloma cell lines and treatments
Five human myeloma cell lines (MM1.S, H929, OPM2, U266, and KMS11) from ATCC-LGC (Molsheim, France). RPMI8226 from ECACC. Cells were maintained in RPMI 1640 medium (Corning Cellgro, Manassas, VA, USA) with L-glutamine supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in a humidified 5% CO2 atmosphere. CX-5641 was obtained from Selleck Chemicals (Houston, USA).
Western blot analysis
5 × 106 cells were obtained by lysing cells with CHIP buffer (1% SDS, 10 nM EDTA, 50 nM Tris HCl (pH 8.1) and protease/ phosphatase inhibitor cocktail) cocktail of protease/phosphatase inhibitors) followed by sonication at 4°C and then assayed using the BC Assay kit (Uptima). A blocking of the specific binding sites was then performed by incubating the membrane in a solution of TBS-T (Tris Buffered Saline Tween: 25 mM Tris, 150 Mm NaCl, 0.05% Tween 20, PH 8.0) containing 1% milk. The membranes were then incubated overnight at 4°C with the following primary antibodies primary antibodies (1:1000) as follows: GAPDH (Santa Cruz Biotechnology, Cat.No.:sc-365062), CASPASE3 (Santa Cruz Biotechnology, Cat.No.: sc-7272), P53 (Cell signaling, Cat.No.:9282), P21 (Chemicon, Car.N. :MAB88058), P27 (Cell signaling, Cat.No.: 2552),PUMA (Cell signaling, Cat.No.:12450), BCL2 (Thermofisher, Cat.No. : 13-8800).
Cell-Titer-Glo viability assay
Human Myeloma Cell Lines (HMCLs) were seeded into 96 well black/clear bottom plate (10.000 cells/well) and exposed to tested drugs for 48 hours. After incubation, cell viability was determined using a 100 μL Cell-Titer-Glo assay (Promega Corp, Madison, WI, USA). The Combination Index (CI) was calculated using Compusyn software.
Cell cycle analysis
MM.1S cells were incubated for 72 hours with CX-5641. Cells were washed with PBS 1X and incubated at 4°C with 70% ethanol for 30 minutes. Cells were washed with PBS and incubated for 20 minutes with PBS-RNASE and 10 minutes with propidium iodide at 4°C. The cell cycle was evaluated by using flow cytometry analysis. Cell cycles were analyzed by using FlowJo VX software.
Immunofluorescence and FISH
MM.1S cell lines were treated with CX-5641 for 2 hours. Cells were fixed with PBS containing 4% paraformaldehyde, permeabilized using PSB containing 0.2% Triton X-100 and then stained using RPA194 antibody (Santa Cruz, sc-48385) and DAPI. ISH cell assay kit (Affymetrix, QVC0001) was used for the FISH experiment. Coverslips were mounted and images were captured with Zeiss Axiovert 200 M inverted microscope with a Plan-Apochromat, 100X/ 1.40 oil DIC x/0.17 objective, and Axio cam MRm camera, and Axiovision software.
RNA extraction, reverse transcription, real-time PCR
Cells were harvested and total RNA was extracted using RNA easy plus mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Whole-cell RNA (250 mg) was converted into cDNA using the Superscript III RT (Life Technologies). Quantitative PCR was carried out using the TaqMan Universal PCR Master Mix and predesigned probes (Life Technologies). RT-PCR for the ITS1 of prerRNA (47S) was accomplished using the SYBR Green PCR Master Mix (Bio-Rad) and the following PCR primer sequences: ITS1,5'TGTCAGGCGTTCTCGTCTC-3'(forward), 5'GAGAGCACG ACGTCACCAC-3'(reverse),GAPDH, 5’TCCCTGAGCTGAACGGG AAG-3’(forward)5’ GGAGGAGTGGGTGTCGCT G-3’ (reverse). All reactions were analyzed using a C1000 Thermal Cycler (Bio-Rad, Hercules, CA, USA). Reactions of pre-designed probes were performed according to the manufacturer's instructions; reactions of the ITS1 and GAPDH were performed as follows: 95°C for 10 min followed by 40 cycles of 95°C for 15 sec. and 62°C for 30 sec. Melting-curve analyses were performed for ITS1 reactions to verify the synthesis of single products. Data were analyzed using the 2-ΔΔCT method.
Statistical analyses
The statistical significance of the data was determined with a Student t-test. P<0.05, P<0.001 and P<0.0001 indicate a statistically significant (*) very significant (**) and highly significant difference (***) respectively.
Results
CX-5461 inhibits rRNA synthesis
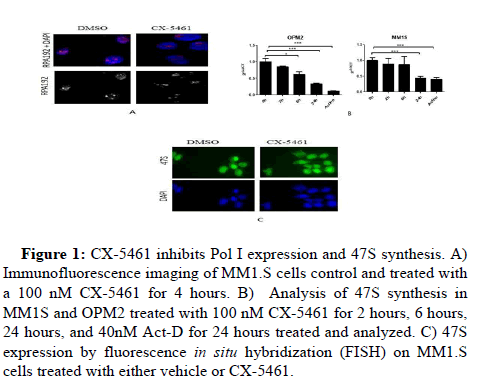
To analyze the effect of CX-5641 in ribosomal stress, we analyzed by IF the level expression of RNA polymerase subunit, RPA194. A decrease of RPA194 was detected after 6 hours exposed to CX-5641 on MM.1S cells (Figure 1A).
Figure 1: CX-5461 inhibits Pol I expression and 47S synthesis. A) Immunofluorescence imaging of MM1.S cells control and treated with a 100 nM CX-5461 for 4 hours. B) Analysis of 47S synthesis in MM1S and OPM2 treated with 100 nM CX-5461 for 2 hours, 6 hours, 24 hours, and 40nM Act-D for 24 hours treated and analyzed. C) 47S expression by fluorescence in situ hybridization (FISH) on MM1.S cells treated with either vehicle or CX-5461.
Since RNA polymerase plays a critical role in the rRNA synthesis, inhibition of Pol I is associates with a decrease of pre-rRNA 47S. To examine the effect of CX-546 on rRNA synthesis, level expression of 47S rRNA were compared in MM.1S and OPM2 cells exposed to DMSO, low dose of Actinomycin D used as control, or CX-5461. It was shown that CX-5461 kinetically suppressed rRNA synthesis. A High inhibition was already shown at 24 hrs. Which suggests that CX-5461 causes a ribosomal disruption (Figure 1B). FISH data confirmed a decrease of 47S on human myeloma cell lines after exposed to CX-5641 (Figure 1C).
CX-5461 triggers p53 pathways and causes cell cycle arrest
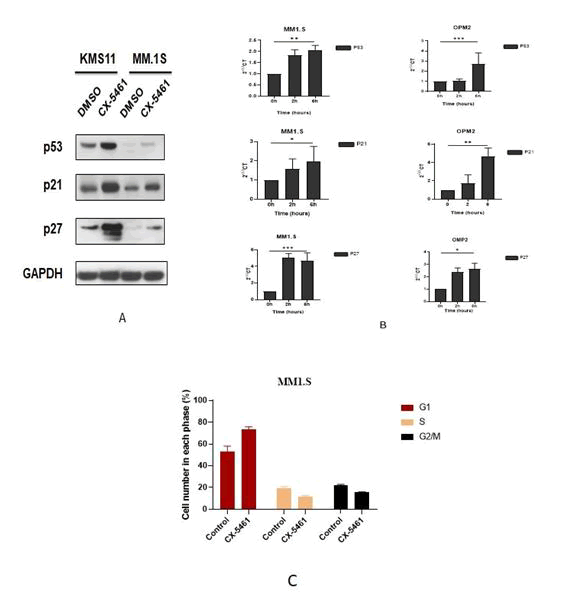
To evaluate the effect of CX-5641 on ribosomal stress, MM.1S, OPM2 and KMS11 cells were treated with CX-5641. An enhancement of p53 and its target p21 and p27 was detected on OPM2 and MM.1S cells (Figure 2A). Kinetic showed that P53, p21 and p27 RNA level were accumulated after 6 hours of treatment in MM.1S and OPM2 cells (Figure 2B). To further investigate the effect of CX-5461 on cell cycle progression. Exposure of cells to 50 nM of CX-5461 significantly arrests the cells in G0/G1‐phase (77.35% ± 9.12%) when compared with control (56.6% ± 3.39%) (Figure 2C).
Figure 2: CX-5461 arrest cycle cells via p53 activation. A) Cells were treated with CX-5461 (100 nM) for 24 hours. The proteins levels: P53,P21 and P27 were analyzed by western blotting. B) Gene expression was measured after treatment with 100 nM CX-5461 for 2 and 6 h in MM1.S and OPM2 cells. Data are expressed as 2-ΔΔct. Results represent the mean ± SD of three independent experiments. Student test was used for statistical analysis (*p<0.05, **p<0.01, ***p<0.001). C) The treatment of 100nM CX-5461 caused a significant increase in the G0/G1 fraction of the cell cycle in MM1S cells.
P21 and P27 were analyzed by western blotting. B) Gene expression was measured after treatment with 100 nM CX-5461 for 2 and 6 h in MM1.S and OPM2 cells. Data are expressed as 2-ΔΔct. Results represent the mean ± SD of three independent experiments. Student test was used for statistical analysis (*p<0.05, **p<0.01, ***p<0.001). C) The treatment of 100nM CX-5461 caused a significant increase in the G0/G1 fraction of the cell cycle in MM1S cells.
CX-5641 down-regulate c-Myc
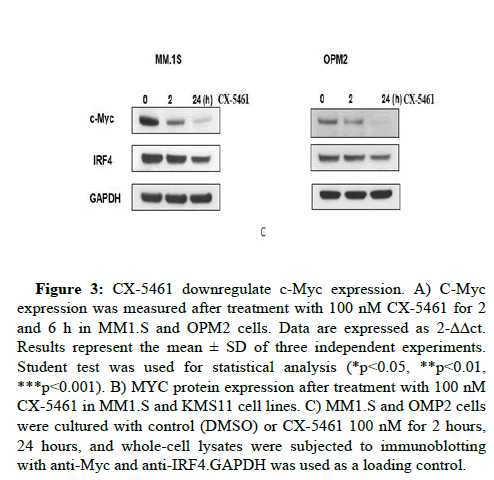
Since p53 down-regulates c-Myc, an accumulation of p53 after exposure to KPT330 is associated with a decrease of c-Myc protein level (Figure 3A). Time course showed that effect was detected after 2 hours of treatment associated with a decrease of the protein expression of IRF4 (Figure 3B). Analysis of c-Myc RNA level showed a significant decrease of c-Myc after 6 hours of exposure to CX-5641 (Figure 3C).
Figure 3: CX-5461 downregulate c-Myc expression. A) C-Myc expression was measured after treatment with 100 nM CX-5461 for 2 and 6 h in MM1.S and OPM2 cells. Data are expressed as 2-ΔΔct. Results represent the mean ± SD of three independent experiments. Student test was used for statistical analysis (*p<0.05, **p<0.01, ***p<0.001). B) MYC protein expression after treatment with 100 nM CX-5461 in MM1.S and KMS11 cell lines. C) MM1.S and OMP2 cells were cultured with control (DMSO) or CX-5461 100 nM for 2 hours, 24 hours, and whole-cell lysates were subjected to immunoblotting with anti-Myc and anti-IRF4.GAPDH was used as a loading control.
CX-5461 is cytotoxic to multiple myeloma cells
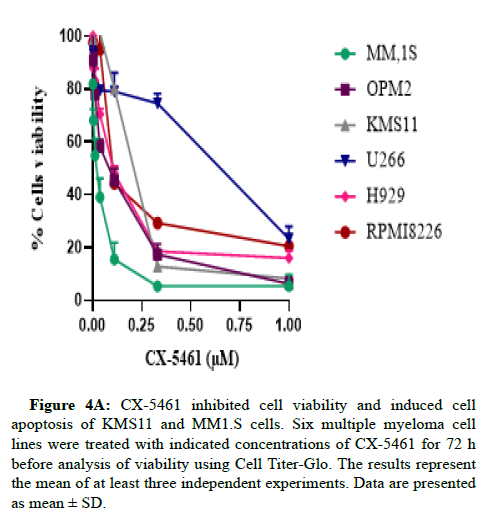
To assess the effect of CX-5461 inhibition in MM cells, we treated human cell lines with different doses of CX-5461 for 72 hr. As shown in Figure 1A, submicromolar concentrations of CX-5461 reduce the viability of MM cells in a dose-dependent manner. The IC50 values ranged from 0.06μM to 0.75 μM (Figure 4A).
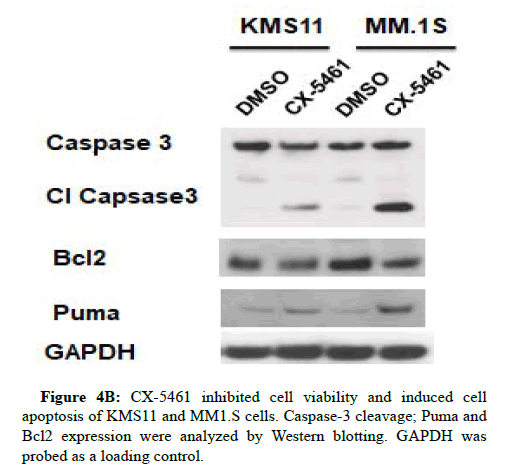
Figure 4A: CX-5461 inhibited cell viability and induced cell apoptosis of KMS11 and MM1.S cells. Six multiple myeloma cell lines were treated with indicated concentrations of CX-5461 for 72 h before analysis of viability using Cell Titer-Glo. The results represent the mean of at least three independent experiments. Data are presented as mean ± SD.
To further investigate apoptosis involved in the CX-5461 stimulus, expression levels of several apoptosis markers were examined. As shown in Figure 4B.
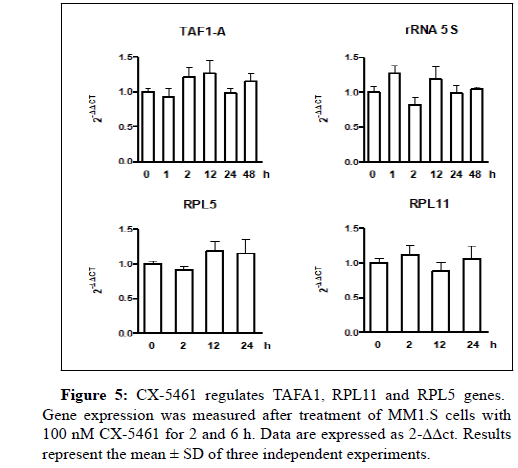
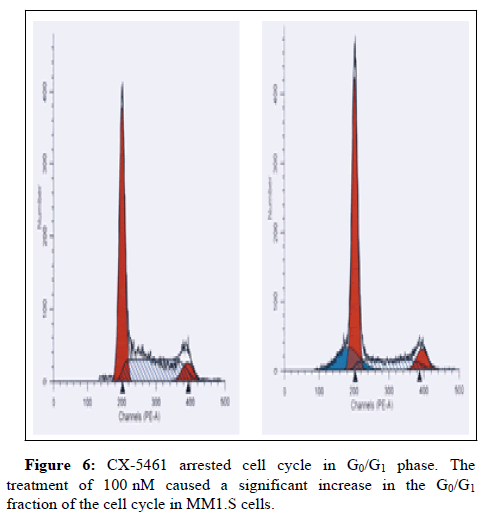
CX-5461 increases the cleavage of caspase‐3 and puma associated with a decrease of Bcl2 protein expression (Figures 5 and 6).
Discussion
The links between ribosome synthesis and cell growth make ribosome biogenesis a key therapeutic target for the treatment of multiple myeloma [9]. Inhibition of ribosome synthesis disrupts cell survival at two levels different: by directly affecting the cell's ability to synthesize proteins, and by indirectly stabilizing the p53 tumor suppressor [10]. It has been shown that up to 80% of cellular energy is consumed in the process of ribosome biogenesis and thus is not surprising, that ribosome biogenesis controls cellular growth and proliferation [11]. Ribosomal biogenesis is a multistep cellular process that involves the processing of pre-ribosomal RNA in the nucleus and the synthesis of ribosomal proteins in the cytosol [12]. The first assembly of ribosomal proteins on the rRNAs is in the nucleus. The final assembly of the subunits 40S and 60S together is in the cytosol [13].
This study we report that CX-5461 inhibits the activity of RNA polymerase I causing the arrest of 47S rRNA synthesis and ribosome biogenesis. Previous studies have shown in vitro and in vivo that the primary target of CX-5461 is the initiation of rDNA transcription by RPI [14]. Our results indicate that CX-5461 inhibits myeloma cell proliferation and induces cell-cycle G1 arrest. Interestingly, we found that CX-5461 led to the up-regulation of p53 and it pathway, associate with the cell cycle arrest and down-regulation of c-Myc. Recent studies have demonstrated that IRF4 is known to upregulate Stage 2, CDK6, and c-Myc and is associated with cellular growth and survival in multiple myeloma [15]. C-Myc promotes the proliferation, cell growth and viability of myeloma cells by the regulation of large number of genes. C-Myc activity is involved in the regulation of ribosomal biogenesis by the regulation of RNA Pol-I. In our study, we confirm that activation of p53 caused by ribosomal stress is associated with a decrease of c-Myc on the RNA level. The downregulation of c- Myc is associated with a decrease of IRF4 and induce of apoptosis. Upon activation, PUMA interacts with anti-apoptotic members of the Bcl-2 family, releasing Bax and/or Bak which are then able to signal apoptosis.
Conclusion
In p53-depend cell cycle arrest is critical for the sensitivity of myeloma cells to CX-5461. Myeloma cells are characterized by a high level of immunoglobulin secretion. This leads to a higher level of ribosomal activity and fibrogenesis [16]. As a consequence, myeloma cell death is efficiently after a ribosomal disruption. Our data suggest that CX-5461 causes a p53-independent nucleolar stress reaction and a nucleolus-specific DNA damage regulation [17]. CX-5461 is involved in the regulation of p53, c-Myc, and ribosomal biogenesis. Blocking the activity of 47S causes a ribosomal disruption associated with an inhibition of c-Myc and an accumulation of p53 signals. Our study reports that CX-5461 is a new therapeutic target in myeloma.
Acknowledgments
The research project was supported by the laboratory of Molecular and Cell Hematology in Pasteur Institute of Tunis. This work was the recipient of financial support from the Tunisian Ministry of Education.
References
- Lee HC (2017) RNA Polymerase I Inhibition with CX-5461 as a Novel Therapeutic Strategy to Target MYC in Multiple Myeloma. Br J Haematol 177:80‑94
[Crossref] [GoogleScholar] [Pubmed]
- Holien T, Misund K, Olsen OE (2015) MYC amplifications in myeloma cell lines: correlation with MYC-inhibitor efficacy. Oncotarget 6:22698-22705.
[Crossref] [Googlescholar] [Pubmed]
- Chiecchio L (2009) Loss of 1p and rearrangement of MYC are associated with progression of smouldering myeloma to myeloma: sequential analysis of a single case. Haematologica 94:1024‑1028.
[Crossref] [Googlescholar] [Pubmed]
- Chanukuppa V (2019) XPO1 is a critical player for bortezomib resistance in multiple myeloma: A quantitative proteomic approach. J Proteomics 209:103-504.
[Crossref] [Googlescholar] [Pubmed]
- Durie (2003) Myeloma management guidelines: a consensus report from the Scientific Advisors of the International Myeloma Foundation. Hematol J 4: 79‑398.
- Madru C, Leulliot N, Lebaron S (2017) La synthèse des ribosomes, au cœur du contrôle de la prolifération cellulaire. Med Sci (Paris) 33:613‑619
- Pecoraro A, Pagano M, Russo G, Russo A, (2021) Ribosome Biogenesis and Cancer: Overview on Ribosomal Proteins. Int J Mol Sci 22:5496
- Turi Z, Lacey M, Mistrik M, Moudry P (2019) Impaired ribosome biogenesis: mechanisms and relevance to cancer and aging (Albany NY) 11 : 2512‑2540
- Nait Slimane S (2020) Ribosome Biogenesis Alterations in Colorectal Cancer. Cells 9:11
- Chaillou T, Kirby TJ, McCarthy JJ (2014) Ribosome biogenesis: emerging evidence for a central role in the regulation of skeletal muscle mass, J Cell Physiol, 229:1584‑1594
- Narla A, Ebert BL (2010) Ribosomopathies: human disorders of ribosome dysfunction. Blood 115:3196‑3205
- Fox JM, Rashford RL, Lindahl L (2019) Co-Assembly of 40S and 60S Ribosomal Proteins in Early Steps of Eukaryotic Ribosome Assembly. Int J Mol Sci 20: 2806
- Pan M (2021) The chemotherapeutic CX-5461 primarily targets TOP2B and exhibits selective activity in high-risk neuroblastoma. Nat Commun 12:64-68
- Butrym A, Łacina P, Rybka J, Chaszczewska-Markowska M, Mazur G, et al. (2016) Cereblon and IRF4 Variants Affect Risk and Response to Treatment in Multiple Myeloma Arch Immunol Ther Exp (Warsz) 64:151‑156, 2016
- Ismael M, Webb R, Ajaz M, Kirkby KJ, Coley HM (2019) The Targeting of RNA Polymerase I Transcription Using CX-5461 in Combination with Radiation Enhances Tumour Cell Killing Effects in Human Solid Cancers. Cancers, 11:10
- James A, Wang Y, Raje H, Rosby R, DiMario P (2014) Nucleolar stress with and without p53. Nucleus 5:402‑426
- Bywater MJ (2012) Inhibition of RNA Polymerase I as a Therapeutic Strategy to Promote Cancer-Specific Activation of p53. Cancer Cell 22:51‑65
Citation: Tagoug A, Novel RNA Polymerase I Inhibitor CX-5461 Exhibits Antitumor Activity in Multiple Myeloma. J Oncol Res Treat 7: 189.
Copyright: © 2022 Tagoug A. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Open Access Journals
Article Usage
- Total views: 1471
- [From(publication date): 0-2022 - Mar 13, 2025]
- Breakdown by view type
- HTML page views: 1212
- PDF downloads: 259