New Electrophoretic Method for Separating Glu and Lys Plasminogen by using DoE Approach
Editor assigned: 01-Jan-1970 / Reviewed: 01-Jan-1970 / Revised: 01-Jan-1970 /
Abstract
Background: Human Plasminogen (PLG, EC 3.4.21.7) is a single chain glycoprotein. This protein circulates in blood as an inactive zymogen. Due to post-translational modification two distinct glycoforms has been found: Glycoform I (GluI-PLG) and Glycoform II (GluII-PLG).A proteolytic activation with physiological relevance consists in the plasmin-catalyzed hydrolysis of the N-terminal amino acid sequence at position 77. This activation can occur on GluI-PLG or GluII-PLG producing respectively LysI-PLG or LysII-PLG. For a pharmaceutical company that produces a drug based on Glu-PLG it is important to develop a method that allows checking the presence of Lys-PLG to exclude some phenomena of pre-activation in the Drug Product.
Study design and methods: A new electrophoretic method to separate the Glu-PLG (I and II) forms from the Lys-PLG (I and II) one was developed. The Design of Experiment (DoE) approach has been employed to set the specific parameters of electrophoresis. The Voltage the Electrophoretic runs time and the loaded protein amount.
Results: Two batches of Plasminogen manufactured by Kedrion were analyzed for their content of Lys-PLG. No trace of this form has been highlighted. The Relative Fronts (RF) of the different protein bands was considered and the elaboration of the DoE results allowed obtaining a robust and reproducible separation of GluI, GluII, LysI and LysII protein band. In addition, the method limit of detection (LOD) was investigated.
Conclusion: The developed method is able to separate the Glu-PLG forms from the Lys-PLG one. The method can be used for checking the stability of a PLG-based drug during shelf life.
Keywords
Glu-Plasminogen;Electrophoresis; DoE approach; Lys-Plasminogen
Introduction
Plasminogen is a very important molecule for the human body [1].Many physiological processes require the activation of Plasminogen to Plasminwhich is essentially regulated by two enzymes: Urokinase and Tissue Plasminogen Activator [2]. The zymogen, full-size form, circulates in the plasmain two distinct Glycoformsdue to post-translational modification. GluI-PLG has an N-linked and O-linked carbohydrate at Asn289 and Thr346, respectively. GluII-PLGcontains only the O-linked [3]. Other Plasminogen forms than the native Glu-PLG have been reported in literature. In particular, a truncated form of Plasminogen which is of physiological importance has been extensively characterized [4].This truncated form originates from a proteolytic digestion carried out by Plasmin which removes the first 77 amino acids from the N-terminal of the protein.In this way the first amino acid of this truncated form turns out to be Lysine, for this reason it is identified as Lys- PLG.Proteolytic cleavage can occur on GluI-PLG or GluII-PLG originating LysI-PLG or LysII-PLG respectively [5,6].
The deficiency of PLG can lead to development of different pathologies. In particular, two types of PLG deficiency have been described: Type I and Type II. Type I is associated to reduced amount of PLG in Plasma, in the other hand Type II is characterized by a normal amount of PLG in circulation [2]. Ligneous conjunctivitis, a rare disease caused by accumulation of fibrin sediments inside the conjunctivae, is the most common form of pathology associated with Type I deficiency [3]. In the treatment of this pathology, topical administration of PLG has been used with good results to minimize the formation of membranes or fibrin sediments [7,8].
A drug for the treatment of ligneous conjunctivitis was developed by KedrionBiopharma purifying Glu-PLG from Human Plasma and used as an orphan drug for the treatment of ligneous conjunctivitis[8]. The importance of a test capable of discriminating between the two forms of Plasminogen (full vs. truncated) is very useful to evaluate the stability of the PLG Drug Product (DP) due to the lower stability of the pre-activated PLG.
No other electrophoretic fast, simple and optimized method capable of separating Glu and Lys-PLG and developed using the DoE approach has been found in the literature.In the past some chromatographic methods have been used to purify and separate the zymogen and the pre-activated forms of the PLG [9,10].For the setting of the experimental conditions it was decided to use a completely innovative approach for the development of such an analytical method: the DoE.The acrylamide gel electrophoresis was chosen for its simplicity of execution and for its potential in separating proteins with different molecular weight.
Materials And Methods
Materials
Bolt 8% Bis-Tris Plus, NuPAGE LDS Sample Buffer 4X, NuPAGE MOPS Running Buffer 20X was supplied by Invitrogen.Lys- plasminogen Standard (Sigma).Simply Blue Safe Stain and Precision Plus Protein™ All Blue Prestained Protein Standards (#1610373) were supplied by Bio-Rad.
Kedrion Plasminogen Standard was manufactured by KedrionBiopharma purifying it fromhuman Plasma in order to use it as orphan drug in the compassionate use.
Methods
Preliminary risk assessment:A risk assessment was performed to assess whether each considered factor couldbe critical in the separation of protein bands associated to full-size or truncated form.
In the choice of the percentage of polyacrylamide gel, a careful research was made. The information provided by the precast gel supplier (Invitrogen) wasanalyzed. It was considered that the maximum separation power, for proteins of such molecular weight (about 89 kDa), was ensured by an 8% polyacrylamide gel.
The choice of carrying out the electrophoretic run in denaturing but not reducing conditions was performed on the basis of previous experiences of the working group. In fact, in previous tests, a more efficient separation of the differentglycoforms of PLG was observed avoiding the addition of reducing reagents. It should be noted that in those electrophoretic conditions the PLG bands have a very different Relative Front (RF) compared to the ones of reducing conditions. Consequently, the molecular weight of the protein is different from that universally recognized (about 89 kDa). Probably, the conformation, obtained by preserving the disulfide bridges of the PLG biochemical structure, increases the resolving capacity of the method.
The choice of using MOPS running buffer instead of MES running buffer (both supplied by Invitrogen) was made on the basis of preliminarytests carried out in the Kedrion R and D laboratories.
It was decided to study thecriticalfactorswith high impact according to the Risk Analysis and their interactions in order to obtain optimal conditions for the separation of the protein bands of PLG, as shown in Table 1.
| Input Factor | Type of impact |
|---|---|
| Voltage | Medium |
| Time of running | High |
| Amount of protein loaded (μg) | High |
| Electrophoresis Run Buffer | High |
| Volume of protein loaded (μl) | Low |
Table 1: Analysis of the factors considered critical for the separation of protein bands in an electrophoresis test.
PLG electrophoretic run
SDS-PAGE was carried out in denaturing conditions using 8% Bis- Tris Plus gel. Glu-PLG (Kedrion internal standard) and Lys-PLG standard were prepared using LDS sample buffer 4x as denaturing agent followed by thermal denaturation at 95°C for 5 minutes.
Theconditions of the critical parameters with high impact were tested according to Table 2.The Standard and Samples were the same for each run: Standard (STD) Glu-PLG, STD Lys-PLG, PLG samples (DP 02LP19A-R and 03LP19A-R) and a mix of STD Glu-PLG/STD Lys-PLG. The spiked sample was prepared by mixing the same amount of Glu and Lys-PLG (1, 2,5 or 4 μg of protein).Once the electrophoretic run was complete, the gel was stainedwith Simply Blue Safe protein stain for 1 h and subsequently de-stained with 5 washing step,one per hour. The gel was acquired withChemiDoc MP using ImageLab software. The molecular weight of each protein band was calculated in reference to molecular weight marker (250, 150, 100, 75, 50, 37 and 25Kda) by ImageLab software. The RF was calculated by ImageLab for all bands ofeachline.
| DoE Matrix | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| StdOrder | RunOrder | A | B | C | ||||||||
| 013 | 1 | 0 | 0 | 0 | ||||||||
| 012 | 2 | 0 | 1 | 1 | A=Voltage (Volt) | |||||||
| 002 | 3 | 1 | -1 | 0 | -1 | 0 | 1 | |||||
| 009 | 4 | 0 | -1 | -1 | 150 | 175 | 200 | |||||
| 011 | 5 | 0 | -1 | 1 | ||||||||
| 006 | 6 | 1 | 0 | -1 | ||||||||
| 005 | 7 | -1 | 0 | -1 | B=Running Time (minutes) | |||||||
| 015 | 8 | 0 | 0 | 0 | -1 | 0 | 1 | |||||
| 003 | 9 | -1 | 1 | 0 | 40 | 50 | 60 | |||||
| 004 | 10 | 1 | 1 | 0 | ||||||||
| 007 | 11 | -1 | 0 | 1 | ||||||||
| 008 | 12 | 1 | 0 | 1 | C=Loaded Protein Amount (µg) | |||||||
| 010 | 13 | 0 | 1 | -1 | -1 | 0 | 1 | |||||
| 014 | 14 | 0 | 0 | 0 | 1 | 2.5 | 4 | |||||
| 001 | 15 | -1 | -1 | 0 | ||||||||
Table 2: DoE Matrixruns order generated by Minitab. The input factors A, B, C were Voltage, Running Time and Amount of loaded protein (µg) respectively. For each input factor 3 different levels were studied 150-175-200 Volt; 40-50-60 minutes and 1-2.5-4 µg
DoE matrix generation
The operating conditions to test the RF proteinperformance were obtained using a three-factor three level Box-Behnken experimental design. The DoE matrix was generated using Software Minitab(version 19.2020.1). In this matrix, a modified central composite design, the variable combinations are at the center and the mid-points of the edges of the variables space. This Experimental Design was preferred respect to other types of matrix because it allowed investigating also the response curvature with a small number of tests. The effect of three independent factors was considered:Voltage (Volt), Running Time (minutes), and loaded Protein Amount(μg). In order to generate the working matrix these factors were named as A, B and C respectively. The matrix setting was reportedin the Table 2.Based on preliminary assessment analysis, for each factor three levels were selected and codified as low (-1), medium (0) and high (+1).
Software Minitab was used for the regression analysis of the data. The significance of independent factors and their interactions were evaluated using the analysis of variance (ANOVA). An alpha (α) level of 0.05 was used to determine the statistical significance in the performed analysis. The optimization of operating conditions to maximize the RF of plasminogen forms were also determined using the Response Optimize function provided by the software.
Results
Electrophoresis run analysis
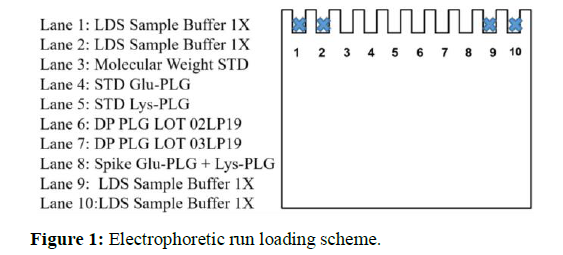
15 runs were performed overall. For each run were analysed: STD Glu-PLG, STD Lys-PLG, PLG samples(DP 02LP19A-R and 03LP19A-R)anda mix of STD Glu-PLG/STD Lys-PLGaccording to the scheme in Figure 1.
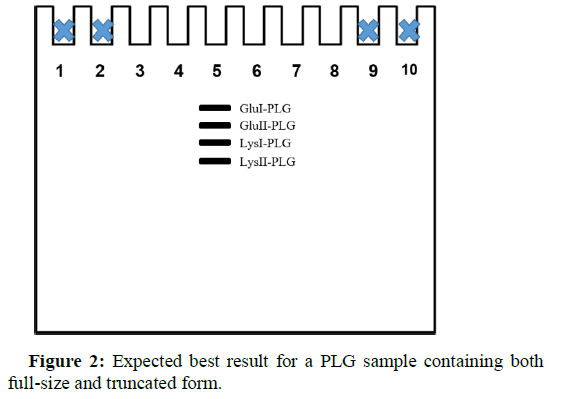
The matrix runs were performed to assign the criticality of each factor (as well as thefactors interactions) on the final result: the best possible separation between the Glu II-PLG and Lys I-PLG bands (Figure 2).
In fact, these two bands are the ones that can probably overlap, making difficult to recognize the four different forms of PLG.Note that Sample buffer (LSD)was loaded in thewells at the ends of the gelin order to avoid “smile effect"(Figure 2).
The tests in the random order were carried outand the results were collected in Table 3. The RF of each protein band was evaluated for each run. The presence of bands attributable to Lys-PLG was not observed in the PLG DP samples.In fact, the analyzed DPs were not out of the shelf-life, consequently no pre-activation of the product was expected.Generally, DPs stored in properly way (in this case<-20°C) were found to be stable, on the contrary DPs subjected to forced degradation showed the presence of degradation bands and Lys-PLG (data not shown).
RunNumber |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | 013 | 012 | 002 | 009 | 011 | 006 | 005 | 015 | 003 | 004 | 007 | 008 | 010 | 014 | 001 |
| Relative Front | |||||||||||||||
| STD Glu-PLG | |||||||||||||||
| Glu I | 0.433 | 0.486 | 0.441 | 0,443 | 0,448 | 0,498 | 0,443 | 0,431 | 0,421 | 0,555 | 0,389 | 0,511 | 0,493 | 0,421 | 0,414 |
| Glu II | 0,454 | 0,513 | 0,464 | 0,462 | 0,471 | 0,523 | 0,463 | 0,455 | 0,445 | 0,586 | 0,413 | 0,542 | 0,519 | 0,446 | 0,439 |
| Lys I | 0.484 | 0.543 | 0.490 | 0.490 | 0.497 | 0.556 | 0.492 | 0.477 | 0.467 | 0.617 | 0.435 | 0.567 | 0.548 | 0.468 | 0.460 |
| Lys II | 0.505 | 0.571 | 0.511 | 0.509 | 0.519 | 0.579 | 0.508 | 0.499 | 0.489 | 0.646 | 0.455 | 0.596 | 0.573 | 0.492 | 0.483 |
| Glu I | 0.440 | 0.490 | 0.445 | 0.445 | 0.453 | 0.503 | 0.448 | 0.435 | 0.426 | 0.558 | 0.393 | 0.514 | 0.494 | 0.423 | 0.419 |
| Glu II | 0.462 | 0.518 | 0.467 | 0.465 | 0.479 | 0.526 | 0.467 | 0.458 | 0.447 | 0.587 | 0.418 | 0.545 | 0.519 | 0.448 | 0.444 |
| Lys I | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Lys II | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Glu I | 0.440 | 0.488 | 0.444 | 0.444 | 0.453 | 0.503 | 0.447 | 0.436 | 0.426 | 0.558 | 0.390 | 0.514 | 0.494 | 0.423 | 0.419 |
| Glu II | 0.463 | 0.516 | 0.465 | 0.462 | 0.479 | 0.526 | 0.467 | 0.460 | 0.449 | 0.587 | 0.417 | 0.546 | 0.519 | 0.448 | 0.444 |
| Lys I | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Lys II | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Glu I | 0.443 | 0.488 | 0.444 | 0.445 | 0.454 | 0.503 | 0.447 | 0.435 | 0.422 | 0.559 | 0.388 | 0.513 | 0.492 | 0.420 | 0.419 |
| Glu II | 0.466 | 0.516 | 0.466 | 0.464 | 0.483 | 0.530 | 0.468 | 0.459 | 0.446 | 0.59 | 0.413 | 0.543 | 0.520 | 0.446 | 0.444 |
| Lys I | 0.489 | 0.547 | 0.488 | 0.49 | 0.503 | 0.56 | 0.495 | 0.489 | 0.475 | 0.627 | 0.441 | 0.580 | 0.553 | 0.475 | 0.467 |
| Lys II | 0.508 | 0.571 | 0.507 | 0.507 | 0.521 | 0.584 | 0.511 | 0.510 | 0.497 | 0.655 | 0.463 | 0.606 | 0.576 | 0.497 | 0.487 |
Table 3: RF Analysis. For each electrophoresis run performed, the RF was calculated by Image Lab software. N/A stays for “not applicable analysis”.
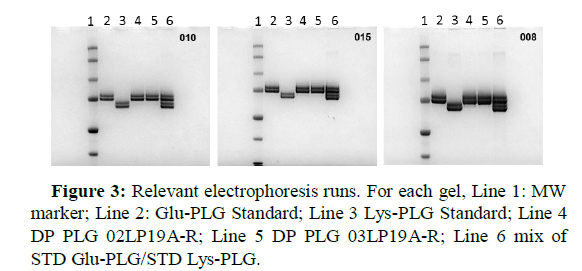
In the spiked prepared samples, four clearly distinct and non- overlapping bands were observed, as shown in Figure 3.Some electrophoretic tests were chosen in order to highlight the effect of the loaded protein amount on the resolution of the protein bands. When 4 μg of Glu-PLG and 4 μg of Lys-PLG (Lane 6, 008 test in Figure 3) were loaded onto the gel, the resolution of the protein bands was not the best possible.Resolution greatly improved when 1 μg of Glu-PLG and 1 μg of Lys-PLG (Lane 6, 010 test in Figure 3) were loaded onto the gel.As consequence,that parameter appeared to be very critical for the desired response from the method.
It should be noted that under the electrophoretic conditions tested, the molecular weight recorded for the PLG was found to be very different from the real one (about 89 kDa), as shown in the Table 4. This phenomenon is due to the different folding that the protein assumes in non-reducing conditions and it wasusedto obtain a better separation between the protein bands.
| Run Number | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | 013 | 012 | 002 | 009 | 011 | 006 | 005 | 015 | 003 | 004 | 007 | 008 | 010 | 014 | 001 |
| MW (Kda) | |||||||||||||||
| STD Glu-PLG | |||||||||||||||
| Glu I | 77.7 | 77.8 | 78.1 | 79.6 | 79.1 | 79.9 | 79.0 | 78.5 | 78.3 | 79.3 | 77.8 | 78.8 | 79.2 | 78.4 | 77.8 |
| Glu II | 74.1 | 74.1 | 74.3 | 75.9 | 74.6 | 76.3 | 75.2 | 74.5 | 74.2 | 75.4 | 73.5 | 74.4 | 75.2 | 74.2 | 73.3 |
| STD Lys-PLG | |||||||||||||||
| Lys I | 70.0 | 70.4 | 70.7 | 71.8 | 70.9 | 72.1 | 71.0 | 71.3 | 71.0 | 72.0 | 70.3 | 71.3 | 71.5 | 71.1 | 70.3 |
| Lys II | 67.1 | 67.2 | 67.8 | 69.2 | 67.8 | 69.3 | 68.7 | 68.3 | 68.0 | 68.9 | 67.3 | 67.9 | 68.6 | 67.9 | 67.1 |
| DP PLG 02LP19A-R | |||||||||||||||
| Glu I | 76.4 | 77.3 | 77.5 | 79.1 | 78.0 | 79.2 | 78.0 | 77.9 | 77.5 | 79.0 | 77.1 | 78.3 | 79.0 | 78.0 | 76.7 |
| Glu II | 73.1 | 73.5 | 73.8 | 75.5 | 73.4 | 75.8 | 74.6 | 74.0 | 73.8 | 75.3 | 72.8 | 74.0 | 75.2 | 73.8 | 72.6 |
| Lys I | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Lys II | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| DP PLG 03LP19A-R | |||||||||||||||
| Glu I | 76.4 | 77.4 | 77.7 | 79.3 | 78.0 | 79.2 | 78.2 | 77.7 | 77.5 | 79.0 | 77.5 | 78.3 | 79.0 | 78.0 | 76.7 |
| Glu II | 73.0 | 73.6 | 74.1 | 75.9 | 73.4 | 75.8 | 74.6 | 73.7 | 73.7 | 75.3 | 73.0 | 73.9 | 75.2 | 73.8 | 72.6 |
| Lys I | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Lys II | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| STD Glu-PLG+Lys-PLG | |||||||||||||||
| Glu I | 76.0 | 77.4 | 77.7 | 79.1 | 77.8 | 79.2 | 78.2 | 77.9 | 78.1 | 78.8 | 78.0 | 78.5 | 79.0 | 78.6 | 76.7 |
| Glu II | 72.5 | 73.6 | 74.0 | 75.7 | 72.8 | 75.3 | 74.4 | 73.9 | 74.0 | 74.9 | 73.5 | 74.3 | 75.0 | 74.2 | 72.6 |
| Lys I | 69.3 | 70.0 | 71.0 | 71.8 | 70.0 | 71.5 | 70.5 | 69.6 | 69.9 | 70.9 | 69.4 | 69.9 | 71.0 | 70.2 | 69.3 |
| Lys II | 66.8 | 67.2 | 68.4 | 69.4 | 67.6 | 68.8 | 68.3 | 66.8 | 67.0 | 68.0 | 66.2 | 66.8 | 68.2 | 67.3 | 66.5 |
Table 4: MW Analysis. For each electrophoresis run, the molecular weight analysis was performed. N/A stay for “not applicable analysis.
DoE matrix output
Lys-PLG forms were not detected in the two batches of Drug Product analyzed. For all tested conditions RF of GluII-PLG was subtracted fromRF of LysI-PLG. This operation was performed for both tested batches: 02LP19A-R and 03LP19A-R. RF difference, as shown in Table 5, was then analyzed using Minitab Software.
| Run Number | Batch 02LP19A-R | Batch 03LP19A-R |
|---|---|---|
| 013 | 0.022 | 0.021 |
| 012 | 0.025 | 0.027 |
| 002 | 0.023 | 0.025 |
| 009 | 0.025 | 0.028 |
| 011 | 0.018 | 0.018 |
| 006 | 0.030 | 0.030 |
| 005 | 0.025 | 0.025 |
| 015 | 0.019 | 0.017 |
| 003 | 0.020 | 0.018 |
| 004 | 0.030 | 0.030 |
| 007 | 0.017 | 0.018 |
| 008 | 0.022 | 0.021 |
| 010 | 0.029 | 0.029 |
| 014 | 0.020 | 0.020 |
| 001 | 0.016 | 0.016 |
Table 5: RF differences (GluII-PLG RF-LysI-PLG RF). This operation was done for all performed runs.
As shown in Table 5, the results recorded for both batches were comparable.
Table 6 shows the quadratic equations describing the relationship between the response and the terms of the model for each batch.
| Batch | Regression Equation in Uncoded Units |
|---|---|
| 02LP19A-R | Y=0,020333+0,003375 A+0,002750 B-0,003375 C+0,000583 A*A+0,001333 B*B+0,002583 C*C+0,000750 A*B-0,000000 A*C+0,000750 B*C |
| 03LP19A-R | Y=0,01933+0,003625 A+0,002125 B-0,003500 C+0,00046 A*A+0,00246 B*B+0,00371 C*C+0,00075 A*B-0,00050 A*C+0,00200 B*C |
Table 6: DoE matrix elaboration. Regression Equation in Uncoded Units of each lot was reported.
These equations were obtained applying the multiple regression analysis on the design matrix, in termsofuncoded units.
In these equations, Y is the response function (RF GluII-PLG RF- LysI-PLG RF) A, B and C are the coded term of the independent factors.
As first step, in this type of analysis it is important to check the reliability of the experimental model considering various parameters. The goodness of fit of the model describes the response(Table 7) and therefore confirmed the answer of the experimental system.
| Batch | S | R-sq | R-sq (adj) |
|---|---|---|---|
| 02LP19A-R | 0,0016683 | 95,22% | 86,61% |
| 03LP19A-R | 0,0025658 | 90,85% | 74,38% |
Table 7: DoE matrix elaboration. Models Summary in Uncoded Units of each lot were reported
For both batches, the small values of S (standard deviation of the distance between the data values and the fitted values) and the high values of regression (0,952 and 0,909) with the relative adjusted regression coefficients (0,866 and 0,744) indicate that the developed quadratic model could adequately describes the system behavior within the selected range of the operating parameters.
As second step, in order to determine whether the association between independent factors (loadedprotein amount, voltage and running time) and the desired response was statistically significant,Analysis of Variance was performed (Table 8). As usually accepted, a significance level of 0.05 was considered as a sufficient requirement.
| Paramaters | Batch 02LP19A-R | Batch 03LP19A-R | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Source | DF | Adj SS | Adj MS | P-Value | F-Value | Adj SS | Adj MS | P-Value | F-Value | |
| Model | 9 | 0.000277 | 0.000031 | 0.008 | 11.06 | 0.000327 | 0.000036 | 0.037 | 5.52 | |
| Linear | 3 | 0.000243 | 0.0000 81 | 0.001 | 29.07 | 0.000239 | 0.000080 | 0.010 | 12.11 | |
| A | 1 | 0.000091 | 0.000091 | 0.002 | 32.74 | 0.000105 | 0.000105 | 0.010 | 15.97 | |
| B | 1 | 0.000061 | 0.000061 | 0.006 | 21.74 | 0.000036 | 0.000036 | 0.066 | 5.49 | |
| C | 1 | 0.000091 | 0.000091 | 0.002 | 32.74 | 0.000098 | 0.000098 | 0.012 | 14.89 | |
| Square | 3 | 0.000030 | 0.000010 | 0.102 | 3.56 | 0.000068 | 0.000023 | 0.108 | 3.46 | |
| A*A | 1 | 0.000001 | 0.000001 | 0.531 | 0.45 | 0.000001 | 0.000001 | 0.745 | 0.12 | |
| B*B | 1 | 0.000007 | 0.000007 | 0.185 | 2.36 | 0.000022 | 0.000022 | 0.125 | 3.39 | |
| C*C | 1 | 0.000025 | 0.000025 | 0.031 | 8.85 | 0.000051 | 0.000051 | 0.039 | 7.71 | |
| 2-Way Interaction | 3 | 0.000005 | 0.000002 | 0.676 | 0.54 | 0.000019 | 0.000006 | 0.474 | 0.97 | |
| A*B | 1 | 0.000002 | 0.000002 | 0.410 | 0.81 | 0.000002 | 0.000002 | 0.584 | 0.34 | |
| A*C | 1 | 0.000000 | 0.000000 | 1.000 | 0.00 | 0.000001 | 0.000001 | 0.713 | 0.15 | |
| B*C | 1 | 0.000002 | 0.000002 | 0.410 | 0.81 | 0.000016 | 0.000016 | 0.180 | 2.43 | |
| Error | 5 | 0.000014 | 0.000003 | N/A | N/A | 0.000033 | 0.000007 | N/A | N/A | |
| Lack-of-Fit | 3 | 0.000009 | 0.000003 | 0.458 | 1.32 | 0.000024 | 0.000008 | 0.368 | 1.87 | |
| Pure Error | 2 | 0.000005 | 0.000002 | N/A | N/A | 0.000009 | 0.000004 | N/A | N/A | |
| Total | 14 | 0.000291 | N/A | N/A | N/A | 0.000360 | N/A | N/A | N/A | |
Table 8: DoE matrix elaboration. Analysis of Variance and relative elaborated parameters, of each batch were reported. In the table N/A stay for Not Applicable. DFThe total degrees of freedom; Adj SS Adjusted sums of squares; Adjusted Adj MS mean squares. P
Results were evaluated with several statistical parameters such as P- Value, F-Value, the degree of freedom (DF), the sum of squares (Adj SS) and the mean sum of the squares (Adj MS).
The significance of each coefficient in equation was determined by means of Fisher’s F-test and P-Value: F>F Critical indicates a significant effect of the factor on the considered response (P<0.05).
Therefore, the larger value of F and the smaller value of P, the more significant is the corresponding coefficient.
As shown in Table 8, all three independent variables have significant effect on the RF GluII-PLG RF-LysI-PLG RFbased on their P-Valuefor both lots DP PLG 02LP19A-R and DP PLG 03LP19A-R (except for B, Running Time, in lot 03LP19).
Also, as evident in Table 8, the quadratic effect of the loaded protein amount was significant on the response function.This important result was alreadyanticipated in Figure 3, where the gels confirming the output of the statistical analysis were shown.
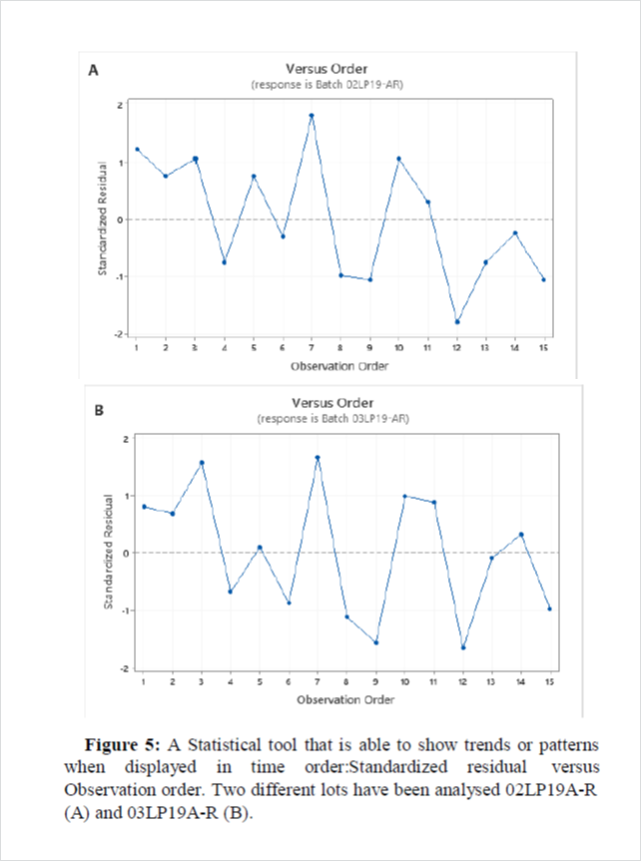
As third step, the normality of the data was evaluated through the normal probability plot of the standardized residual (Figure 4) as well as their distribution respect to the order of execution of the tests (Figure 5). The points on the plot lie on a straight line, the residuals are normally distributed as confirmed by P>0.05. Moreover,independent residuals showed no trends or patterns when displayed in time order. Ideally, the residuals on the plot should fall randomly around the center line. As evident in Figure5, the points are scattered randomly between the outlier detection limits -3 and +3.
Response optimization
The goal of this analysis was find the best run condition to maximize the separation between the bands (GluII and LysIPL Gglycoforms). The optimization consists in setting the values of the analyzed parameters in order to maximize the separation. Minitab software was used to reach this result (Table 9).
| Response | Goal | Lower | Target | Weigt | Importance | |
|---|---|---|---|---|---|---|
| Batch 02LP19A-R | Maximum | 0,016 | 0,030 | 1 | 1 | |
| Batch 03LP19A-R | Maximum | 0,016 | 0,030 | 1 | 1 | |
Table 9: Variable settings to calculate the fits for the responses.
In order to obtain the best operating conditions to maximize the RF difference Lys I-PLG vs Glu II-PLG, the equation is considered as the objective function. The optimization was carried out by means of the numerical technique built in the Minitab Software that searches the design space to achieve the goal of optimization in the range of independent factors. The results showed that factor A (Voltage) was optimized at level +1 (200V), factor B (time) at level +1 (60 min) and factor C (Amount of protein loaded) at level -1 (1μg) for both lots 02LP19-AR and 03LP19-AR (Table 6). The Multiple Response Prediction was reported in the Table 10.
| Impact factor | Levels | Optimized | Optimized values | ||
|---|---|---|---|---|---|
| A (Voltage-V) | -1 (150) | 0 (175) | 1 (200) | 1 | 200 |
| B (Time-minutes) | -1 (40) | 0 (50) | 1 (60) | 1 | 60 |
| C (Loaded Amount-μg) | -1 (1) | 0 (2,5) | 1 (4) | -1 | 1 |
Table 10: Final choice of conditions to be adopted in order to have the best result.
The obtained optimal operating conditions were used in another experimental run to validate the model prediction. The RF obtained experimentally, confirms the reliability of the model (Data not show).
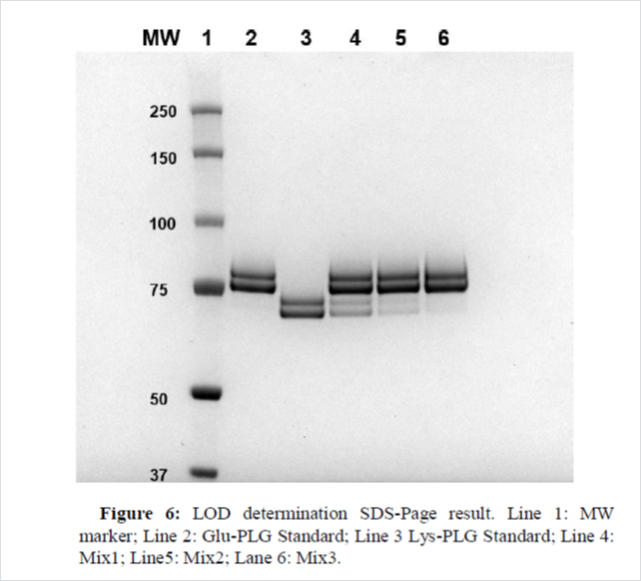
LOD determination
In SDS-Page method, LOD refers to the minimum amount of protein capable of producing a band detectable by the instrument (ChemiDoc).For this parameter three different analytical sessions were performed. In particular, keeping the amount of STD Glu-PLG constant, different amounts of STD Lys-PLG were evaluated in order to observe the disappearance of its band, as consequence, for each session, three different mix were prepared, shown in Table 11.
| Standard | Mix 1 | Mix 2 | Mix 3 |
|---|---|---|---|
| Glu-PLG | 1 µg | 1 µg | 1 µg |
| Lys-PLG | 0.30 µg | 0.10 µg | 0.03 µg |
Table 11: Composition of Glu-PLG and Lys-PLG used for LOD determination
All sessionsprovidedcomparableresultsas shown in Figure 6 (first session). The two bands of the Lys-PLG were clearly visible and distinct when 0.3 μg of Standard was loaded onto the gel (Lane 4). The two bands of Lys-PLG werestill visible, especially theLysII-PLG one, when 0.1 μg of Standard was loaded on the gel, but in this case the instrument used (ChemiDoc) was not able to detect them (Figure 6, Lane 5), while a feebleband was observed when 0.03 μg of Lys- PLG was loaded onto the gel (Figure 6, Lane 6).
Discussion
The Plasminogen/Plasmin system performs several fundamental physiological functions [11,12].Consequently, plasminogen deficiencies can have serious consequences [2].Ligneous conjunctivitis is a pathology characterized by the formation of pseudo membranes, rich in fibrin, in the area of the conjunctiva and is associated with Type I Plasminogen deficiency [13].A pharmaceutical product based on Glu-Plasminogen with great therapeutic potential was manufactured by Kedrion Spa and supplied as a compassionate use for ligneous conjunctivitis.It is very important checking that PLG drug product does not undergo any kind of pre-activation/ degradationduring the shelf life.Therefore, it is necessary to have a testthat can highlight somepre-activations of Glu-PLG to Lys-PLG or Plasmin.
The requirement to separate two different forms of PLG, the full- size (Glu-PLG) form and the truncated one (Lys-PLG), leadstheKedrion R and D laboratoriesto develop a new electrophoretic method.The request was to develop a simple, fast and cheapmethod. A complication was the presence of two glycoforms both for the full-size form (GluI-PLG and GluII-PLG) and for the truncated one (LysI-PLG and LysII-PLG) (8).The major difficulty was in separating the GluII- PLG bandfromtheLysI-PLGoneas a consequence of a similar electrophoretic mobility. The maximization of the distance between the protein bands and in particular between the two aforementioned was the aim of the development of the analytical method.Moreover, without an adequate electrophoretic technique, there is the risk of underestimating the presence of Lys-PLG in a PLG-based drug.
The practice of using the DoE approach in developing an analytical method is spreading very fast. Indeed, this approach offers numerous advantages in evaluating which inputs (or factors) have a significant impact on the robustness of a method and/or the desirable output. Some crucial components of a DoE are: factors, levels and response (output). A preliminary analysis must be conducted in order to have basic information as starting point (i.e. the choice of the type of gel and the percentage of components that form it). 8% acrylamide/bis acrylamide gel was used, as analyzing the migration pattern of the protein band, this type of gel guaranteed a good separation in the range of molecular weights of our interest (66-97 kDa). The choice of the running buffer (MOPS) was made as well on the basis of previous experiences. Finally, in preliminary experiments the possibility of carrying out the electrophoretic run in denaturing and non-reducing conditions wasalready evaluated. This condition proved to be more efficient in the separation of the bands (data not shown), this phenomenon essentially is due to the presence of disulfide bridges in the protein structure of the PLG.
Conclusion
Based on this assumption, themultifactorialanalysis should identify the factors whose interaction could cause the method variability. In the case of the electrophoretic method discussed in this paper, the following factors were considered: the Voltage, the electrophoretic running time and the loaded protein amount. On the basis of previous experiences in the execution of the technique, the levels of investigation for each factor were chosen.For each factor analyzed, three levels were considered: low (-1), middle (0) and high (+1). At the end of this preliminary phase, a DoE matrix was generated using the Minitab software.The 15 tests were performed in order to identify the best conditions and to obtain the desired response: maximizing the separation between the protein bands.
According to thedefinedapproach,it was feasible to develop and optimize a simple and fast method to highlight the presence of Lys- PLG in PLG samples.Finally, with the determination of the LOD of the method; it was possible to clarify the minimum amount of Lys- PLG that can be observed when such a test is carried out.
From the results obtained it can be concluded that the developed method is reliable and reproducible.It will be part of an analytical panel to check the stability of drug products in order to ensure the efficacy and safety of the drug.This new approach to method development, even the simplest, can provide significant benefits in terms of reliability.
Author Contributions
Conceptualization, A.S.; A.C. E.A.; A.F.; methodology, A.S., A.C.,E.A.; investigationA.S., M.C., Z.R; data treatment, A.C. M.C, L.S., Z.R.; writing original draft preparation, A.S.; writing-review and editing, A.F.; E.A. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang L, Seiffert D, Fowler BJ, Jenkins GR, Thinnes TC, et al. (2002) Plasminogen has a broad extrahepatic distribution. Thromb Haemost. 87(3): 493-501.
- Mehta R, Shapiro AD (2008) Plasminogen deficiency. Hemophilia. 14(6): 1261-1268.
- Schuster V, Hugle B, Tefs K (2007) Plasminogen deficiency. J Thromb Haemost. 5(12): 2315-2322.
- Lijnen HR, Van Hoef B, De Cock F, Collen D (1989) The mechanism of plasminogen activation and fibrin dissolution by single chain Urokinase-Type plasminogen activator in a plasma milieu In Vitro. Blood. 73(7): 1864-1872.
- James C, Nesheim ME (1992) Lys-plasminogen is a significant intermediate in the activation of glu-plasminogen during fibrinolysis in Vitro. J Biol Chem. 267(36): 26150-26156.
- Gong Y, Kim SO, Felez J, Grella DK, Castellino FJ (2001) Conversion of glu-plasminogen to lys-plasminogen is necessary for optimal stimulation of plasminogen activation on the endothelial cell surface. J Biol Chem. 276(22): 19078-19083.
- WattsP, Suresh P, Mezer E, Ells A, Albisetti M, et al. (2002) Effective treatment of ligneous conjunctivitis with topical plasminogen. May Am J Ophthalmol. 133(4): 451-455.
- Nardini C (2012) Plasminogen: Its role in the therapy of ligneous conjunctivitis. Production of Plasma Proteins for Therapeutic Use. 311-320.
- Summaria L, Spitz F, Arzadon L, Boreisha IG, Robbins KC (1976) Isolation and characterization of the affinity chromatography forms of human glu-and lys-plasminogens and plasmins. J Biol Chem. 251(12): 3693-3699.
- Gonzalez-gronow M, Violand BN, Castellino FJ (1977) Purification and some properties of the glu-and lys-human plasmin heavy chains. J Biol Chem. 252(7): 2175-2177.
- Aisina RB, Mukhametova LI (2014) Structure and function of plasminogen/plasmin system. Russian J Bioorganic Chem. 40: 590-605.
- Pepper MS. (2001) Role of the Matrix metalloproteinase and plasminogen activator-plasmin systems in angiogenesis. Arterioscler Thromb Vasc Biol. 21: 1104-1117.
- Schuster V, Seregard S. (2003) Ligneous conjunctivitis. SurvOphthalmol. 48(4): 369-388.
Citation: Salvatorea A, Carluccia A, Carforaa ML, Raiaa Z, Sanguignoa L, et al. (2021) New Electrophoretic Method for Separating Glu and Lys Plasminogen by using DoE Approach. J Anal Bioanal Tech 12: 430.
Copyright: © 2021Salvatorea A. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Open Access Journals
Article Usage
- Total views: 1837
- [From(publication date): 0-2021 - Apr 26, 2025]
- Breakdown by view type
- HTML page views: 1112
- PDF downloads: 725