Multivariate Analysis of Agronomically Important Traits of Sunflower [Helianthus Annuus L] Genotypes under Rainfed Condition
Received: 02-Jun-2024 / Manuscript No. acst-24-138938 / Editor assigned: 05-Jun-2024 / PreQC No. acst-24-138938 / Reviewed: 19-Jun-2024 / QC No. acst-24-138938 / Revised: 23-Jun-2024 / Manuscript No. acst-24-138938 / Published Date: 30-Jun-2024
Abstract
A study on the genetic diversity of sunflower genotypes is useful in any breeding program for providing efficient variety development. The oil is considered to be of supreme quality. Sunflower is grown for oil as an edible vegetable oil and industrial purposes in the world. It can also contribute a big share in improving local edible oil production in Ethiopia. But the country has immense potential and suitable land. Sunflower genotypes were evaluated for important quantitative traits using alpha lattice design with two replications at Holeta during the 2020/2021 main cropping season to determine the genetic variability of sunflower genotypes using Multivariate Analysis of agronomically important traits. Significant variation was observed for the studied traits of the genotypes. Principal component analysis showed that the first five principal components out of 16 accounted for 74.5 % of the total variability. So, the genotypes were grouped into five genetically divergent clusters. The 1st principal component which accounted for 38.23% of total variability among sunflower genotypes mainly originated from all characters except oil content. Similarly, the PC2, PC3, PC4 and PC5 which accounted nearly for ~12.0%, ~9.35%, 8% and ~7% respectively. Two hundred twenty sunflower genotypes were grouped into five clusters with different traits. Out of two hundred twenty genotypes, 39.09% was grouped in cluster III followed by 29.09%, 19.09%, 9.54%, and 3.18% for cluster-I, V, II, and IV, respectively. Inter-cluster distances were highly significant but intra-cluster distances were non-significant compared to the chi-square value. The maximum and minimum inter-cluster distance was D2= 2023.6 & D2= 427.46 between cluster-I & II and Cluster-4 & 5 respectively. So, we can conclude that the genotypes are genetically divergent. In sunflower improvement early to medium maturing, a big head & tough stem, seed yield & number per head, medium height, hundred seed weight, oil content, seed filling percentage, head angle & shape, and reaction to stress should be emphasized traits to fill the gap of sunflower productivity through the targeted breeding approach.
Keywords
Cluster; Distances, Divergent, Agronomically-important; Principal component
Introduction
Sunflower (Helianthus annuus L., 2n=34) is an important oil crop and belongs to the family Asteraceae (Compositae). The genus, Helianthus comprises 49 species and 19 subspecies with 12 annual and 37 perennial herbaceous species (Balogh, 2008). Its oil is highly used in the human diet because of its high level of unsaturated fatty acids, lack of linolenic acid, and bland flavor (Putnam et al., 1990). Its oil may also be used as a raw material to extract bio-diesel which can be used as fuel in diesel engines (Antolin et al., 2002; Murugesan et al., 2009) [1].
The Production (5150.5t), productivity (1.03t/ha), and area (4749.96ha) coverage in Ethiopia are low and below the world average (2.14 t/ha) (FAOSTAT, 2022). But the country has immense potential and suitable land. Since sunflower can better fit in both main season and irrigated cropping systems, and have high oil contents, tolerance to drought, and a high yield potential; the country can exploit the benefits of sunflower but not yet. Due to limited number of improved varieties and different constraints of current agricultural policy issues; like “Ten in Ten”; and the edible oil imported from abroad, the country is unable to produce and use widely [2].
In Ethiopia, Sunflower research began by introducing germplasms from the USA, Russia, Canada, Yugoslavia Romania, and Italy to develop varieties with high oil content and stable performance in different Agroecology (Hiruy, 1990). Getting uniform variety in terms of agronomic important traits, height, and head, besides low yield potential is the main challenge in sunflower breeding. This is mainly due to cross-pollination and the limited number of germplasms in the research system. To overcome those challenges, the presence of basic information on genetic variability is important to plan breeding strategies and make effective selection or crossing for desirable traits. Variations are very precious for any breeding program (Allard, 1961; Hussain, 2015) as success mainly relies upon the extent of variation present in a germplasm for yield and its contributing traits (Qamar, 2015).
Genotypic variations existing in the genotypes can be exploited efficiently when genetic resources in breeding programs exist (Sowmya, 2010). Thus, during the selection or crossing program starts for traits of interest more consideration has to be given to those attributes with low environmental variability. Yield is a complex trait and is collectively influenced by various component traits that are polygenically inherited (Allard, 1999; Falconer & Mackay, 1983).
Plant breeders can apply multivariate analysis tool to investigate clear patterns of variability existing among genotypes. The results of multivariate analysis are of greater benefit to identifying the parents for improving various traits and it can also be exploited in the planning and execution of future breeding programs (Mustafa, 2015). It also offers an opportunity for the exploitation of appropriate genotypes for developing specific plant traits (Pecetti & Damania, 1996) [3]. Additionally, (Nazir, 2013) stated that Multivariate analysis is a reliable tool for the successful selection of parents in breeding programs. Keeping in view the importance of this technique, researchers used it in sunflower genetic variability studies. So far, there are minimal reports on studies done to assess the level of genetic variability available in sunflower germplasm in Ethiopia except a few studies made involving a limited number of genotypes, realizing the importance of the crop and the existing information gap in the present study was done to investigate the PCA, Cluster Analysis and genetic divergence of genotypes based on agronomically important traits for future exploitation [4].
Materials and Methods
Description of Study Areas
A description of the study area is presented in (Table 1) (Figure 1).
| Descriptor | Location |
|---|---|
| Holeta | |
| Distance from Addis Ababa | 32 |
| Latitude | 9º 00’ N |
| Longitude | 38º30’ E |
| Altitude (m a. s. l.) | 2400 |
| Mean annual rainfall (mm) | 1144 |
| Mean annual temperature (°C) | Min=6°C |
| Max.=22°C | |
| Soil type | Nitosols |
| Soil drainage | Well drained |
| Soil pH | 5.2-6.0 |
| Organic C (%) | 1.18 |
| N (%) | 0.16 |
| Source: Haile M. and Mola T., 2019 | |
Table 1: Description of the study area
Experimental Design and Trial Management
The experiment was conducted at Holeta Agriculture Research Center during the 2020/21 main cropping season. The experiment was arranged in an 11x20 Alpha Lattice design with two replications. Each genotype was planted having 0.25m spacing between plants, 0.75m spacing between rows, and 1.5m row length. Recommended fertilizer was applied at the rate of 23/23 kg NP2O5 kg per hectare. Other agronomic and management practices were applied uniformly as per the recommendation for the area. Yield and yield-related data were collected from randomly selected and tagged plants. Seed yields were adjusted to a moisture content of 7% using grain moisture analysis [5].
Data collected
The following data were collected for quantitative traits by using Descriptors of the International Board for Plant Genetic Resources (IBPGR, 1985) for sunflower. Number of Leaves per plant, LNPP; Plant height (PH) in cm, Head Diameter (HD) in cm; Petiole length (PL) in cm; Days to 50% flowering (DF); Days to maturity (DM); Stem Diameter (SD) in cm, Seed filling percentage (SFP); Hundred Seed weight (HSWT) in g, Seed yield per plant (SYPP) in g; Percent oil content (OC); Seed yield per hectare (SYPH) in kg; Oil yield (OYPH) in kg/ha; and Percent stand [6].
Statistical data analysis
Multivariate analysis is useful for the characterization and classification of genotypes evaluated for agronomically important traits. In this study, principal component analysis (PCA), Cluster Analysis, and Genetic divergence were computed. The data were standardized to a mean zero and variance of unity before computing PCA to avoid scale differences used for recording data on different traits (Sneath and Sokal, 1973) [7].
Principal component analysis was computed by using Minitab v17 (Minitab, 1998) and also confirmed by SAS 9.3 computer software (SAS, 2014). Clustering of genotypes was done using the PROC clustering strategy of SAS 9.3 and Minitab v-17 but appropriate numbers of clusters were determined by Eigenvalue greater than 1 using Minitab v-17 software. Inter and intra-cluster genetic distance was calculated using Mahalanobis's (Mahalanobis, 1936) generalized distance statistical formula, and the values calculated between pairs of clusters were considered as chi-square values and tested for significance using p-degrees of freedom, where “p” indicates the number of traits used.
〖D^2〗_ij=(X_i-X_j)S^(-1) (X_i-X_j)
Whereas; Dij2 is the distance between groups i and j; Xi and Xj are the vectors of the means of the traits for groups i and j and S-1 is the pooled within groups variance-covariance matrix [8].
Result and Discussion
Principal component analysis
Principal component (PC) analysis showed only the first five principal components out of sixteen traits account for most of the variability extracted since five principal components had Eigenvalue greater than one. They account for 74.5 % of the total variability in the original data. The 1st principal component which accounted for 38.23% of total variability among genotypes mainly originated from all characters except oil content. Similarly, the 2nd principal component (PC2) which accounted nearly for 12.0% of the total variability among genotypes was attributed to discriminatory traits of Days to flowering, days to maturity, plant height, stem diameter, leaf number per plant, and stand percentage. The other way, the highest negative contribution in PC2 was from head diameter, petiole, yield per plot, yield per head, oil content, oil yield per hectare, seed filling percentage, hundred seed weight, seed number per plant, and yield per hectare (Table 2) [9].
PC |
Eigenvalue | % Total - variance | Cumulative - Eigenvalue | Cumulative - % |
|---|---|---|---|---|
| 1 | 6.12 | 38.23 | 6.12 | 38.23 |
| 2 | 1.92 | 11.98 | 8.03 | 50.21 |
| 3 | 1.50 | 9.35 | 9.53 | 59.56 |
| 4 | 1.28 | 8.00 | 10.81 | 67.56 |
| 5 | 1.11 | 6.92 | 11.92 | 74.49 |
Table 2: Eigenvalues of correlation matrix, and related statistical values
The 3rd principal component which contributed 9.3% of total variability among sunflower genotypes was from discriminatory traits mainly, days to flowering, days to maturity, yield per plot, oil yield per hectare, and yield per hectare. However negative principal contributors in PC3 are plant height, stem diameter, head diameter, leaf number per plant, petiole length, stand percentage, yield per head, oil content, seed filling percentage, hundred seed weight, and seed number per plant. The 4th principal component which contributed 8.0% to the total variability among sunflower genotypes was because of plant height, stem diameter, head diameter, leaf number per plant, petiole length, yield per plot, oil content, oil yield per hectare and seed yield per hectare [10]. Conversely, the negative contribution was from days to flowering, days to maturity, stand percentage, yield per head, seed filling percentage, hundred seed weight, and seed number per plant. Finally, the 5th principal component contributed nearly 7.0% to the total variability. Out of sixteen traits considered days to flowering, days to maturity, plant height, leaf number per plant, petiole length, stand percentage, oil content, and oil yield per hectare were positive contributors. Stem diameter, head diameter, yield per plot, yield per head, seed filling percentage, hundred seed weight, seed number per plant, and yield per hectare contributed negatively. Out of sixteen quantitative traits considered days to flowering, days to maturity, plant height, leaf number per plant, and oil yield per hectare contributed positively to the variation among genotypes in four principal components out of the five principal components. Unfortunately, seed number per plant, hundred seed weight, and seed filling percentage contributed positively not only in PC1 but for the rest of PCs negatively [11].
Different scholars have done principal component analysis for their respective sunflower genetic variability studies. According to Safavi et al. (2011) applied principal component analysis in their cultivar development program and stated the potential use of principal component analysis in the selection of superior genotypes in sunflower. Kholghi et al. (2011) also applied principal component analysis for the estimation of genetic diversity in sunflower. Nazir et al. (2013) reviewed numerous yield-related characters and reported the contribution of the first two components in the assessment of the total variation of sunflower hybrids [12]. Mohammed (2019) also reported the 1st principal component contributed 31.9 % of total variability mainly originating from characters of yield per plant, head diameter, stem diameter, Ray floret number, yield per plant, number of seeds per plant, yield per hectare, head diameter, stem diameter, plant height, days to flowering, days to maturity and seed filling percentage. Also, PC2 which accounted for 22.72 % of the total variability attributed to discriminatory characters of yield per plant, yield per hectare, oil yield per hectare, seed filling percentage, and hundred seed weight. The 3rd, 4th, and 5th PC contributed 12.25%, 10.11 %, and 1.08 % of the total variability were attributed to discriminatory characters respectively. Similarly, out of the thirteen traits, four principal components exhibited more than 1 Eigenvalue and showed about 80.2% variability among the genotypes studied. PC1 had the highest variability (41%) followed by PC2 (17.9%), PC3, (12.6%), and PC4 (8.7%) for traits (Table 3) [13].
| Traits | Eigenvectors | ||||
|---|---|---|---|---|---|
| PC1 | PC2 | PC3 | PC4 | PC5 | |
| DF (days) | 0.201 | 0.542 | 0.146 | -0.133 | 0.029 |
| DM (days) | 0.225 | 0.519 | 0.104 | -0.121 | 0.091 |
| PH (cm) | 0.336 | 0.222 | -0.044 | 0.033 | 0.135 |
| SD (cm) | 0.327 | 0.067 | -0.161 | 0.191 | -0.086 |
| HD (cm) | 0.165 | -0.100 | -0.321 | 0.557 | -0.054 |
| LNPP (No) | 0.228 | 0.102 | -0.122 | 0.225 | 0.257 |
| PL (cm) | 0.292 | -0.033 | -0.200 | 0.272 | 0.027 |
| Stand (%) | 0.097 | 0.109 | -0.412 | -0.300 | 0.444 |
| Yld/P (g) | 0.346 | -0.169 | 0.320 | 0.010 | -0.097 |
| Yld/H (g) | 0.299 | -0.219 | -0.145 | -0.315 | -0.147 |
| OC (%) | -0.053 | -0.305 | -0.015 | 0.018 | 0.716 |
| OY/ha (Kg) | 0.312 | -0.285 | 0.316 | 0.030 | 0.171 |
| SFP (%) | 0.082 | -0.135 | -0.146 | -0.423 | -0.030 |
| HSWT (g) | 0.084 | -0.047 | -0.503 | -0.079 | -0.331 |
| SNPP (No) | 0.269 | -0.221 | -0.111 | -0.346 | -0.071 |
| Yld/ha (kg) | 0.346 | -0.169 | 0.320 | 0.010 | -0.097 |
| Eigenvalue | 6.1168 | 1.9164 | 1.4917 | 1.2751 | 1.1134 |
| Proportion | 0.382 | 0.120 | 0.093 | 0.080 | 0.070 |
| Cumulative | 0.382 | 0.502 | 0.595 | 0.675 | 0.745 |
Table 3: Eigenvectors Eigenvalues, proportion, and cumulative variance for five PCs
principal component analysis depends on findings of which variables are most strongly correlated with each component, i.e., which of these values are large in magnitude, the farthest from zero in either direction influence the clustering more than those with lower values closer to zero (Chahal & Gosal, 2002). As per Kline (2014), the contribution of 0.30 or higher can be considered important. So, the maximum contribution in PC1 was by yield per hectare and yield per plot followed by plant height, stem diameter, and oil yield are important. Whereas, in PC2 days to flowering and days to maturity are the maximum contributor. In PC3 maximum contributions were by yield per hectare, yield per plot, and oil yield per hectare. The maximum contribution in PC4 was by head diameter. In PC5 maximum contribution was by oil content followed by stand percentage [14].
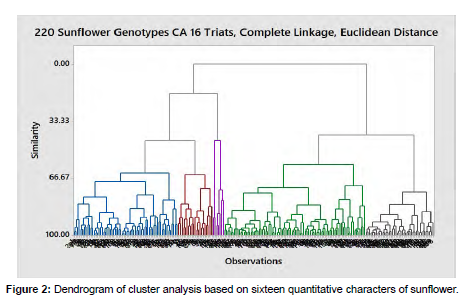
Cluster analysis
Sunflower genotypes were grouped using Euclidean distances of dissimilarity to their distinct groups. The D2 values based on the means of genotypes resulted in categorizing the 220 sunflower genotypes into five clusters based on the result of PC Analysis having eigenvalue ≥1 which depicts the existence of genetic divergence among genotypes. Accordingly, Cluster-I consists of 64 genotypes (29.09%); Cluster-II consists of 21 genotypes (9.54%); Cluster-III has 86 genotypes (39.09%), Cluster-IV consists of 7 genotypes (3.18%) and Cluster-V consisted of 42 genotypes (19.09%) of the total studied population (Table 4) [15].
Cluster No. |
Number of genotypes | Proportions % | The average distance from Centroids | Maximum distance from Centroids |
|---|---|---|---|---|
| I | 64 | 29.09 | 564.98 | 1121.54 |
| II | 21 | 9.54 | 653.34 | 914.03 |
| III | 86 | 39.09 | 558.12 | 1282.81 |
| IV | 7 | 3.18 | 1004.67 | 1549.35 |
| V | 42 | 19.09 | 320.13 | 799.00 |
Table 4: Cluster number, Number of Genotypes, Proportion, Average distance from Centroids, and Maximum distance from Centroids
Sunflower genotypes included under Cluster-I were characterized by late flowering and maturity, tall plants with tough stems, nearly medium-headed, leafy with short petiole length, medium stand percentage, medium yield per head, and oil content with high oil yield, medium seed filling percentage, medium hundred seed weight, medium seed number per plant and high seed yield per hectare compared to the grand mean of genotypes. Therefore, selection based on days to flowering, maturity, and plant height could not be advisable from cluster-I genotypes for developing early flowering and maturity and short to medium variety [16]. The genotypes in this cluster had high seed yield and oil yield, medium oil content, seed filling percentage, hundred seed weight, seed number per plant, head diameter, and seed yield per head. Therefore, selection for genotypes late in flowering and maturity, high seed yielder, tough stem with medium headed and oil content is recommended from genotypes of this cluster. Cluster-II was characterized by late flowering and maturity, tall plants with tough stems, big-headed once, leafy with long petiole, good stand percentage, high yield per head and plot, nearly low oil content, high oil yield with high seed filling percentage, low hundred seed weight, high seed number per plant head and seed yield per hectare as well compared to the grand mean of genotypes. This depicts that selection can be made for head diameter, tough stem, seed yield, and also a selection for late flowering and maturing once, for tall plants as well from genotypes of this cluster (Figure 2) [17].
Genotypes included under Cluster-III have characters of medium in flowering and maturity period, intermediate height with thin stem, nearly medium headed, medium leaf number with short petiole length, nearly medium stand percentage, nearly good seed yield per plot and seed yield per head, medium oil content and oil yield per hectare, intermediate seed filling percentage, medium hundred seed weight, nearly intermediate seed number per plant head and intermediate seed yield per hectare compared to grand mean of genotypes. So, Selection for early flowering, maturity, short to medium plant height, medium head diameter, medium oil content, seed filling percentage, and seed number per plant with high hundred seed weight from genotypes of this cluster can be an opportunity to improve sunflower genotypes [18].
Similarly, Cluster-IV included genotypes can be characterized by a medium in flowering and late maturing period, tall plants with tough stems, big-headed, medium leaf number and petiole length, excellent stand percentage, seed yield per plot and seed yield per head, high oil content and oil yield per hectare, intermediate seed filling percentage and hundred seed weight, high seed number per plant head and intermediate seed yield per hectare compared to the grand mean of genotypes. Selection for early maturing, tough stem with big-headed, high oil content, hundred seed weight, and high seed yield per head and per hectare and also high oil yield per hectare from genotypes of this cluster can be an opportunity to improve sunflower genotypes in those characters. Also, Cluster-V consists of genotypes that have traits of early flowering and maturity periods, short plants with tin stem, nearly medium headed, low leaf number with short petiole length, intermediate stand percentage, low seed yield per plot, and seed yield per head, high oil content and low oil yield per hectare, low to intermediate seed filling percentage and hundred seed weight, low seed number per plant head and intermediate seed yield per hectare compared to grand mean of genotypes. This implies that selection for these traits from genotypes of this cluster can be effective in the improvement of sunflower genotypes through selection (Table 5).
| Variable | Cluster | Centroid Mean |
||||
|---|---|---|---|---|---|---|
| I | II | III | IV | V | ||
| DF (days) | 110.74 | 115.45 | 105.54 | 106.00 | 98.17 | 106.61 |
| DM (days) | 164.81 | 175.17 | 156.79 | 163.50 | 142.69 | 158.40 |
| PH (cm) | 210.30 | 235.67 | 179.06 | 200.14 | 139.52 | 186.67 |
| SD (cm) | 2.28 | 2.62 | 1.99 | 2.29 | 1.57 | 2.06 |
| HD (cm) | 15.40 | 16.02 | 15.09 | 16.89 | 13.28 | 14.98 |
| LNPP (No) | 43.40 | 47.10 | 39.96 | 40.46 | 34.86 | 40.69 |
| PL (cm) | 14.39 | 15.89 | 12.85 | 13.79 | 10.42 | 13.15 |
| Stand (%) | 77.99 | 79.37 | 76.26 | 86.90 | 76.98 | 77.54 |
| Yld/P (g) | 341.07 | 465.97 | 196.32 | 291.56 | 72.99 | 243.65 |
| Yld/H (g) | 84.95 | 149.60 | 66.28 | 171.95 | 32.48 | 76.57 |
| OC (%) | 26.96 | 25.64 | 27.01 | 28.74 | 28.47 | 27.20 |
| OY/ha (Kg) | 818.05 | 1059.29 | 468.88 | 743.07 | 182.48 | 580.86 |
| SFP (%) | 92.37 | 94.53 | 92.58 | 92.94 | 91.29 | 92.47 |
| HSWT (g) | 6.19 | 5.87 | 6.19 | 6.43 | 5.466 | 6.03 |
| SNPP (No) | 1265.23 | 2102.6 | 1100.95 | 3150.64 | 665.655 | 1356.47 |
| Yld/ha (kg) | 3031.69 | 4141.92 | 1745.04 | 2591.64 | 648.792 | 2165.79 |
Table 5: Cluster means for sixteen quantitative characters of sunflower genotypes
Genetic divergence
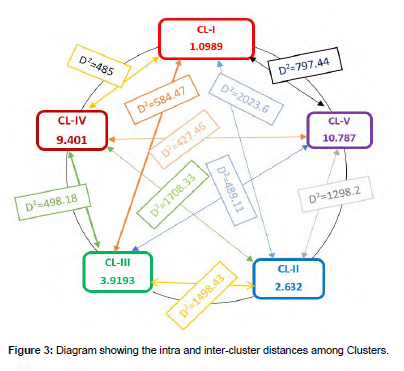
Pair-wise generalized distances (D2) among the five clusters are presented below. The genetic distance analysis showed that all inter-cluster distances were highly significant while there was a non-significant intra-cluster distance. The maximum genetic distance was observed between cluster-I and cluster-II (D2=2023.6) followed by cluster-II and cluster-IV (D2=1708.33), cluster-II and cluster-III (D2=1498.43) and cluster-II and cluster-V (D2=1298.2), most divergent clusters. These high values of inter-cluster distances may result due to the different genetic backgrounds of the genotypes. The least genetic distance was found between cluster-IV and cluster-V (D2=427.46) followed by cluster-I and cluster-IV (D2=485.0) and between cluster-III and cluster-V (D2=489.11) [19].
The X2=test for the five clusters revealed highly significant inter-cluster distance for all clusters and non-significant Intra-cluster distance between genotypes. This shows that there is considerable genetic variability in studied genotypes. So, clusters with the largest genetic distance indicate the real variability between the genotypes of those clusters. A unique and maximum variation is expected from crosses of parents from clusters with high inter-cluster distance. Thus, crossing sunflower genotypes selected from cluster-I and cluster-II; cluster-II and cluster-IV, cluster-II and cluster-III, cluster-I and cluster-III, and those from cluster-III and cluster-IV could be a source of desirable variation as the inter-cluster distance between them were large respectively. But, depending on the specific objectives of the breeding program genotype selection should consider the advantages of each cluster and each genotype in that cluster. So, according to the results of the study genotype with the highest oil content found in Custer-IV should be considered if the breeding objective is to develop high-oil-content sunflower genotypes. Suppose your breeding objective is to improve the seed (achene) yield of the sunflower genotype considering cluster-II genotypes which showed the highest mean value for seed yield per hectare [20].
According to Keneni et al. (2005) and Ghaderi et al. (1984), a crossing of genotypes from distant inter-clusters may produce blessing heterosis at F1 generations and a wide range of variability in subsequent segregating populations and this is a great opportunity for a breeder to get this result. Tyagi et al. (2013) studied genetic variability among 18 sunflower inbred lines, and the genotypes were grouped into five clusters based on D2 analysis. Shamshad et al. (2014) studied using 31 germplasm lines of sunflower for yield and yield-related traits and grouped them into six clusters based on D2 analysis indicating the presence of a high level of diversity in the genetic material (Table 6) (Figure 3) [21].
Cluster |
1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 | 1.0989NS | 2023.6** | 584.47** | 485** | 797.44** |
| 2 | 2.6322NS | 1498.43** | 1708.33** | 1298.2** | |
| 3 | 3.9193NS | 498.18** | 489.11** | ||
| 4 | 9.401NS | 427.46** | |||
| 5 | 10.787NS |
Table 6: Inter-cluster (off-diagonal) and Intra-cluster (diagonal) distance between clusters
Conclusion and Recommendation
For any genetic improvement program for target traits, the choice of parent is important, and traits of interest should be taken into consideration for component breeding to obtain an appropriate ideotype plant. The results of multivariate analysis show that the first five principal components were found to be significant and accounted for about 74.49% of the gross genetic variability among genotypes. Each principal component; PC1, PC2, PC3, PC4, and PC5 contributed 38.2%, 12%, 9.3%, 8.0%, and 7% to the gross genetic variation of the genotypes respectively. Two hundred twenty sunflower genotypes were grouped into five clusters with different traits like plant height, stem diameter, yield, days to flowering and maturity, oil yield and content, head diameter, and stand percentage. Out of two hundred twenty genotypes, 39.09% was grouped in cluster-III followed by 29.09%, 19.09%, 9.54%, and 3.18% for cluster-I, V, II, and IV, respectively. Inter-cluster distances were highly significant but intra-cluster distances were non-significant compared to the chi-square value at 15 degrees of freedom. The maximum and minimum inter-cluster distance was D2= 2023.6 & D2= 427.46 between cluster-I & II and Cluster-4 & 5 respectively.
So, we can conclude that based on sixteen studied quantitative traits the 220 sunflower genotypes are genetically divergent. Genotypes from divergent clusters, such as, between cluster-I & II, cluster-II & III, cluster-II & IV, and cluster-II & V are recommended sources of parental material for sunflower improvement through hybridization. Crossing among genotypes of those clusters selected for specific traits of interest may help bring a new gene pool and expand the range of adaptation. Continuous selection in advance generation may lead to the development of lines with high yield combining desirable component traits. Improvement of sunflower genotypes having traits namely early to medium maturing, big-headed & tough stem, seed yield & number of seeds per head, medium plant height, hundred seed weight, oil content, seed filling percentage, head angle & shape, and reaction to biotic and abiotic stress should be emphasized to obtain genotypes with high seed yield and oil content for the farming society and investors as well.
Acknowledgments
The author would like to express his thanks to Holeta Agriculture Research Centre (HARC) for Providing all research facilities and laboratory services to carry out this research work.
Conflict of Interest
The authors declare no conflict of interest.
Data availability statement: All the data are available.
References
- Allard RW (1961) Principles of plant breeding. Soil Science 91: 414.
- Antolın G, Tinaut FV, Briceno Y, Castano V, Perez C, et al. (2002) Optimisation of biodiesel production by sunflower oil transesterification. Bioresource technology 83: 111-114.
- Balogh, L. (2008) Sunflower Species. Helianthus spp.) In: Botta-Dukát, Z. & Balogh, L.(eds.). The most important invasive plants in Hungary 227-255.
- Chahal GS, Gosal SS (2002) Principles and procedures of plant breeding: Biotechnological and conventional approaches. Alpha Science Int'l Ltd.
- Ghaderi A, Adams MW, Nassib AM (1984) Relationship between genetic distance and heterosis for yield and morphological traits in dry edible bean and faba bean. Crop Sci 24: 37-42.
- Haile M, Mola T (2019) Seed coating; Genius Coat Cereal Disco AG L-439; effect on seed yield and yield related traits of malting barley (Hordeu mvulgare L.) in Central Highlands of Ethiopia. Inter J Agri Biosci, 8: 141-147.
- Hussain F, Iqbal J, Rafiq M, Mehmood Z, Ali M, Ramzan N, et al. (2015) GENETIC IMPROVEMENT IN LINSEED (LINUM USITATISSIMUM L.) THROUGH VARIABILITY HERITABILITY AND GENETIC ADVANCE. J Agric Res 53(4).
- Keneni G, Jarso M, Wolabu T, Dino G (2005) Extent and pattern of genetic diversity for morpho-agronomic traits in Ethiopian highland pulse landraces: I. Field pea (Pisum sativum L.). Genetic Resources and Crop Evolution 52: 539-549.
- Kholghi M, Bernousi I, Darvishzadeh R, Pirzad A (2011) Correlation and path-cofficient analysis of seed yield and yield related trait in Iranian confectionery sunflower populations. African Journal of Biotechnology 10: 13058-13063.
- Lagiso TM, Singh BCS, Weyessa B (2021) Evaluation of sunflower (Helianthus annuus L.) genotypes for quantitative traits and character association of seed yield and yield components at Oromia region, Ethiopia. Euphytica 217:1-18.
- Mahalanobis PC (1936) On the generalized distance in statistics. Proceedings of National Institute of Sciences. India 2: 49-55.
- Murugesan A, Umarani C, Subramanian R, Nedunchezhian N (2009) Bio-diesel as an alternative fuel for diesel engines-a review. Renewable and sustainable energy reviews 13: 653-662.
- Mustafa HSB, Jehanzeb F, Ejaz H, Tahira B, Tariq M, et al. (2015) Cluster and principal component analyses of maize accessions under normal and water stress conditions. J of Agric Sciences 60: 33-48.
- Nazir A, Farooq J, Mahmood A, Shahid M, Riaz M, et al. (2013) Estimation of genetic diversity for CLCuV, earliness and fiber quality traits using various statistical procedures in different crosses of Gossypium hirsutum L 43(4).
- Pecetti L, Damania AB (1996) Geographic variation in tetraploid wheat (Triticum turgidum spp. Turgidum convar. Durum) landraces from two provinces in Ethiopia. Genetic Resources and Crop Evolution 43: 395-407.
- Putnam DH, Oplinger ES, Hicks DR, Durgan BR (1990) Sun flower alternative field crop manual. Coop extension service wiscons. USA.
- Qamar R, Sadaqat HA, Bibi A, Tahir MHN (2015) Estimation of combining abilities for early maturity, yield and oil related traits in sunflower (Helianthus annuus L.). Int J Sci Nature 6: 110 -114.
- Safavi AS, Safavi SM, Safavi SA (2011) Genetic Variability of some morphological traits in Sunflower (Helianthus annuus L.). Am J Scient Res 17: 19-24.
- Shamshad M, Dhillon SK, Tyagi V, Akhatar J (2014) Assessment of genetic diversity in sunflower (Helianthus annuus L.) germplasm. Int J Agric Food Sci Tech 5: 824-827.
- Sowmya HC, Shadakshari YG, Pranesh KJ, Srivastava A, Nandini B, et al. (2010) Character association and path analysis in sunflower (Helianthus annuus L.). Ele J Plant Breed 1: 828-831.
- Tyagi V, Dhillon SK, Bajaj RK, Kaur J (2013) Divergence and association studies in sunflower (Helianthus annuus L.). Helia, 36:77-94.
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Citation: Mola T (2024) Multivariate Analysis of Agronomically Important Traits ofSunflower [Helianthus Annuus L] Genotypes under Rainfed Condition. Adv CropSci Tech 12: 706.
Copyright: © 2024 Mola T. This is an open-access article distributed under theterms of the Creative Commons Attribution License, which permits unrestricteduse, distribution, and reproduction in any medium, provided the original author andsource are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 1997
- [From(publication date): 0-2024 - Nov 05, 2025]
- Breakdown by view type
- HTML page views: 1698
- PDF downloads: 299