Multivariate Analysis among Advanced Breeding Lines of Rice (Oryza Sativa L.) Under Sub-Tropical Ecology of Jammu Region of J&K
Received: 01-Nov-2022 / Manuscript No. jpgb-22-79696 / Editor assigned: 04-Nov-2022 / PreQC No. jpgb-22-79696 (PQ) / Reviewed: 18-Nov-2022 / QC No. jpgb-22-79696 / Revised: 25-Nov-2022 / Manuscript No. jpgb-22-79696 (R) / Published Date: 30-Nov-2022
Abstract
Present study was aimed to determine the nature and extent of genetic divergence for agro-morphological and quality traits among twenty seven advanced breeding lines of rice utilizing multivariate techniques. Based on the calculated D2 values, breeding lines were grouped into six non overlapping clusters. Inter-cluster distances were found to be relatively greater than intra-cluster distances indicating presence of enormous amount of variation among the lines. Cluster I and Cluster II were found to be the largest clusters consisting of 14 and 9 genotypes respectively, while, remaining four clusters were monogenotypic consisting of single line only. Maximum inter-cluster distance was reported between cluster II and VI, while maximum intra-cluster distance was recorded for cluster II respectively [1]. Breeding lines present in cluster II were observed to have maximum cluster means for traits viz., plant height, and total number of tillers per plant, amylose content, grain yield per plant and grain yield per plot indicating its superiority over rest of the clusters. Traits including grain yield per plot, grain yield per plant, days to maturity, 1000 grain weight, and total number of tillers per plant and panicle length were observed to be major contributors to genetic divergence. Principal component analysis revealed presence of genetic variation through six principal components with first five components viz., PC-I to PC-IV having eigen scores greater than one suggesting that traits within these axis have potential effect on development of a phenotype. Further principal component analysis revealed that traits such as number of effective tillers per plant, grain yield per plot, plant height, panicle length and days to 50 per cent flowering contribute maximum towards genetic diversity [2].
Keywords
Rice breeding lines; Clusters and principal component analysis
Introduction
Rice (Oryza sativa L.) is a major and vital cereal grain, next only to wheat in terms of global cultivated area. It is the staple food of more than half of the population of world. In addition to calories, rice is a good source of magnesium, phosphorus, manganese, selenium, iron, folic acid, thiamine and niacin; but it is low in fibre and fat (Fukagawa and Ziska, 2019). In India rice is cultivated over an area of about 45.07 million hectares in India with production and productivity of 122.27 million tons and 2.71 tons per hectare respectively (Anonymous, 2022), while in Union Territory of Jammu and Kashmir it was cultivated over an area of 267.58 thousand hectares with production and productivity of 5186 thousand quintals and 21.74 quintals per hectare respectively (Anonymous, 2022). Rice is one among few cereal crops with large amount of genetic variation and is being cultivated from sea level to higher elevations. The availability of significant genetic diversity for key characteristics in the parents being hybridized is the most important pre-requisite for starting a successful breeding operation which in turn helps in realizing the true production potential for developing improved rice varieties adapted to various agro-climatic zones [3-5].
A quantitative assessment of genetic diversity directs breeders towards speedy progress in breeding programs to get preferred recombinants in segregating generations. Hybrids with high heterosis are typically derived from parental genotypes differing in common ancestry, ecotype, and geographic origin and so on. Morphological, biochemical and DNA markers can be used to assess genetic diversity within breeding populations however; traditionally it is assessed using the D2 approach based on morphological features [6]. This approach gives a measure of the proportional contribution of different traits to diversity both within and across clusters and genotypes selected from highly divergent clusters is likely to yield heterotic pairings and considerable heterogeneity in successive generations. D2 statistics allows separation of different advanced breeding lines and provide a good picture regarding the diversity of breeding material. Tocher's method reveals the clustering pattern and provides measure of genotype separation inside and between the clusters. The extent of heterogeneity among advanced breeding lines belonging to different clusters is determined by cluster means for various agro-morphological and quality traits [7-9]. Present study endeavors to quantify the presence of genetic diversity among 27 advanced breeding lines of rice for agromorphological and quality traits using D2statistics (Mahalanobis, 1928).
Materials and Methods
Twenty seven advanced/stable breeding lines of rice available with Division of Plant Breeding and Genetics were evaluated for agromorphological and quality traits so as to assess diversity among them for exploiting in hybridization programme. Experimental material was evaluated in Randomized Complete Block Design with three replications at Research Farm of the Division of Plant Breeding and Genetics, Faculty of Agriculture, SKUAST-J, Main Campus, Chatha, Jammu during kharif 2020 [10]. The weather during kharif season (June to November) was ideal and supportive for optimum crop growth. Average rainfall was recorded was 866.6mm while, minimum and maximum temperature ranged from 9.20°C (November) to 36.90°C (March, June). All the recommended cultural practices were followed as per schedule to provide favorable environment for crop growth. Observations on different agro-morphological and quality characteristics viz., plant height (cm), total number of tillers per plant, number of effective tillers per plant, panicle length (cm), 1000 grain weight(g) and grain yield per plant (g) were recorded on five randomly chosen plants from each replication to identify the type and degree of genetic divergence in the experimental material, while as some of observations viz., days to 50 per cent flowering (no), days to maturity (no) and grain yield per plot (kg) were taken on per plot basis [11]. Grains of each line were dehulled for grain quality assessment such as length and breadth of kernel, length-breadth ratio and amylose content respectively. Genetic diversity among advanced breeding lines of rice was determined by employing the D2 statistic given by Mahalanobis in the year 1936 and principal component analysis was determined by method explained by Harman (1976) [12] .
Results and discussions
Analysis of variance (Table 1) revealed presence of significant differences among advanced breeding lines of all the agromorphological and quality traits viz., days to 50 per cent flowering, plant height, total number of tillers per plant, number of effective tillers per plant, days to maturity, panicle length, 1000 grain weight, kernel length, kernel breadth, length-breadth ratio, amylose, grain yield per plant and grain yield per plot. The presenceof enormous amount of genetic variation variability within advanced breeding lines may be due to inherent differences in the parents being hybridized. Sarwar et al., 2015; Konate et al., 2016 and Dhakal et al., 2021 findings are in agreement with the findings of the present study [13].
| Source | Degrees of freedom | Days to 50% | PH (cm) | Total number of tillers per plant | Number of effective tillers per plant | DTM | PL (cm) | 1000 grain weight (g) | Kernel | Kernel | L/B | Amylose content | Grain yield per plant | Grain yield per plot |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| of | flowering (no) | length (mm) | breadth (mm) | ratio | (%) | (g) | (kg) | |||||||
| variation | ||||||||||||||
| Replication | 2 | 174.16 | 197.05 | 9.32 | 5.69 | 812.3 | 17.4 | 60.15 | 16.75 | 0.37 | 2.3 | 0.8 | 33.38 | 0.3 |
| Breeding lines | 26 | 140.73** | 370.99** | 33.67** | 25.14** | 131.37** | 22.36** | 12.88** | 5.74** | 0.22** | 0.85* | 58.80** | 41.67** | 0.36** |
| Error | 52 | 37.23 | 21.82 | 4.6 | 3.78 | 43.38 | 3.97 | 4.1 | 2.03 | 0.09 | 0.46 | 0.08 | 9.02 | 0.07 |
** and * represents significance level at 1 % and 5 %
* PH: Plant height, DTM: Days to Maturity, PL:Panicle length
Table 1:Analysis of variance for agro-morphological and quality traits in advanced breeding lines of rice.
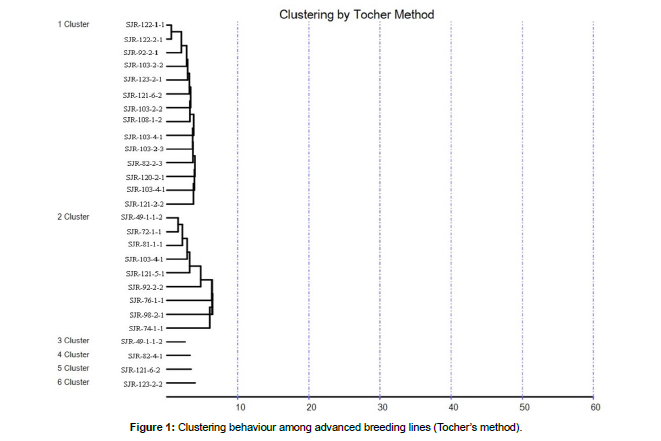
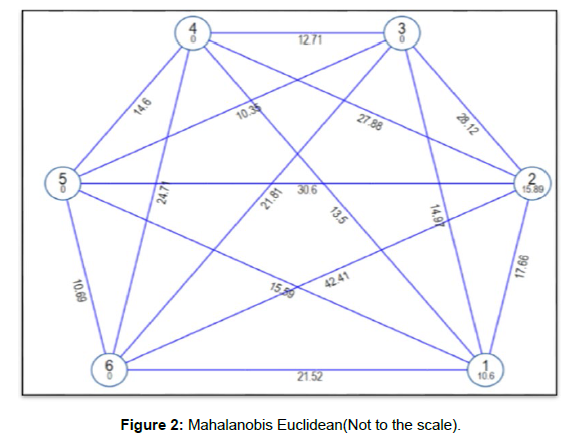
On the basis of clustering pattern as revealed by Tocher’s method (Rao, 1952) advanced breeding lines were categorized into six (6) non overlapping clusters (Table 2 and Figure 1) indicating presence of enormous amount genetic diversity among advanced breeding lines of rice, similar clustering pattern was reported by (Manohara et al., 2013; Dubey et al., 2018 and Ranjith et al., 2018). Tocher's approach was used to arrange the advanced breeding linesinto six clusters based on average D2 values [14]. Further perusal of experimental results reported that out of six clusters, the first two clusters were multi-genotypic consisting of fourteen (14) and nine (9) genotypes respectively, while rest four clusters were mono-genotypic consisting of single genotype only. The maximum number of advanced breeding lines were reported in clusters (I and II), because most these lines are sister lines of each other as depicted by their pedigree record. The advanced breeding lines came from single source and were dispersed through multiple clusters indicating the presence of significant amount diversity within the genotypes. The diversity among the advanced breeding lines of common origin may arise due to factors like heterogeneity, genetic architecture and earlier selection for specific trait [15]. The maximum inter-cluster distances (Table 3 & Figure 2 ) were reported between cluster II and cluster VI (42.41), indicating presence of highly diverse advanced breeding lines in both the clusters, while as minimum intercluster distance was recorded between cluster III and cluster V (10.35) indicating genotypes in these clusters are closely related. The maximum intra-cluster distance was reported for cluster II (15.98) indicating the presence of significant amount diversity among the genotypes in this cluster. The advanced breeding lines of rice belonging to most diverse clusters based on intra and inter cluster distances can be utilized as parents in hybridization programmes to get wide range of variability, transgressive segregants and desirable heterotic combinations. A similar recommendation was proposed by (Murthy and Arunachalam, 1996 and Rahman, 1997) [16]. Cluster means (Table 4) for various agromorphological and quality traits revealed the presence of enormous amount of the genetic variability for all the traits. The maximum cluster mean for characters viz., plant height (127.36), total number of tillers per plant (15.44), grain yield per plant (24.18) and grain yield per plot (1.78) were reported in cluster II, reflecting its superiority over rest of the clusters. Further perusal of experimental findings confirmed that traits viz., grain yield per plot, grain yield per plant, 1000 grain weight, days to maturity, total number of tillers per plant and panicle length contributed maximum toward genetic divergence (Table 5). The contribution of these traits in total towards genetic divergence is 84.92 %. The traits contributing maximum towards genetic divergence (D2 value) need to be given top priority during selection and hybridization programmes (De et al., 1988).
| Cluster No. | No. of advanced breeding lines | Description of advanced breeding lines |
|---|---|---|
| I | 14 | SJR-103-2-2, SJR-103-2-2, SJR-103-4-1, SJR-103-4-1, SJR-108-1-2, |
| SJR-120-2-1, SJR-121-2-2, SJR-121-6-1, SJR-122-1-1, SJR-122-2-1, | ||
| SJR-123-2-1, SJR-103-2-3, SJR-92-2-1, SJR-82-2-3 | ||
| II | 9 | SJR92-2-2, SJR-98-2-1, SJR-121-5-1, SJR-49-1-1-2, SJR-81-1-1, SJR-76-1-1, SJR-74-1-1, SJR-102-4-1, SJR-72-1-1 |
| III | 1 | SJR-49-1-1-2 |
| IV | 1 | SJR-82-4-1 |
| V | 1 | SJR-121-6-2 |
| VI | 1 | SJR-123-2-2 |
Table 2: Allocation of experimental material in various clusters.
| Cluster | I | II | III | IV | V | VI |
|---|---|---|---|---|---|---|
| I | 10.6 | 17.66 | 14.97 | 13.5 | 15.59 | 21.52 |
| II | 15.89 | 28.12 | 27.88 | 30.6 | 42.41 | |
| III | 0 | 12.71 | 10.35 | 21.81 | ||
| IV | 0 | 14.6 | 24.71 | |||
| V | 0 | 10.69 | ||||
| VI | 0 |
Table 3: Intra (diagonal) and inter cluster distances.
| Cluster No. | Days to 50% | PH | Total No. of | No. of | DTM | PL (cm) | 1000 | Kernel length | Kernel breadth | L/B | Amylose | Grain | Grain |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| flowering | (cm) | tillers | effective | grain weight | (mm) | (mm) | ratio | (%) | yield per | yield per | |||
| per plant | tillers per plant | (g) | plant | plot | |||||||||
| (g) | (kg) | ||||||||||||
| Cluster I | 93.83 | 113.86 | 12.54 | 11.22 | 133.07 | 27.26 | 27.54 | 10.06 | 2.31 | 6.03 | 19.05 | 23.18 | 1.68 |
| Cluster 2 | 92.78 | 127.36 | 15.44 | 13.41 | 135 | 28.35 | 28.25 | 9.58 | 2.21 | 5.85 | 24.02 | 24.18 | 1.78 |
| Cluster 3 | 85.67 | 100.56 | 8.4 | 6.33 | 131.67 | 22.77 | 22.77 | 9.61 | 1.84 | 5.58 | 22.31 | 18.79 | 1.25 |
| Cluster 4 | 106.33 | 104.59 | 11.27 | 10.27 | 142.67 | 29.33 | 23.33 | 10.04 | 2.05 | 6.04 | 19.23 | 21.74 | 1.65 |
| Cluster 5 | 91 | 97.02 | 13.53 | 12.37 | 125 | 25.3 | 28.27 | 11 | 2 | 6.5 | 21.25 | 18.55 | 1.73 |
| Cluster 6 | 95 | 94.75 | 12.8 | 14.33 | 123 | 23.21 | 28.53 | 8.6 | 2.23 | 5.42 | 20.01 | 22.15 | 1.69 |
* PH: Plant height, DTM: Days to Maturity, PL:Panicle length
Table 4: Cluster means among agro-morphological and quality traits.
| Agro-morphological and quality Agro-morphological and quality traits |
Times ranked 1st | Contribution towards |
|---|---|---|
| genetic divergence | ||
| Days to 50 % flowering | 1 | 0.28% |
| PH (cm) | 11 | 1.98% |
| Total number of tillers per plant | 35 | 9.97% |
| Number of effective tillers per plant | 5 | 1.42% |
| DTM | 36 | 10.26% |
| PL (cm) | 29 | 8.26% |
| 1000 grain weight (g) | 55 | 15.67% |
| Kernel length (mm) | 18 | 5.13% |
| Kernel Breadth (mm) | 17 | 4.84% |
| L/B ratio | 8 | 2.28% |
| Amylose (%) | 6 | 1.15% |
| Grain yield per plant (g) | 63 | 17.95% |
| Grain yield per plot (kg) | 75 | 20.81% |
* PH: Plant height, DTM: Days to Maturity, PL:Panicle length
Table 5: Contribution by individual trait towards total genetic divergence.
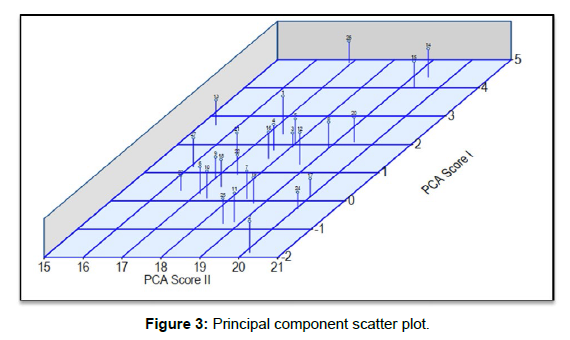
Principal component or canonical root analysis revealed that the genetic divergence in advanced breeding lines of rice and cumulative variance of about 81.24 per cent (Table 6 & Figure 3) as depicted by the first five vectors with Eigen value greater than one, suggesting that identified agro-morphological and quality traits with in the axis demonstrated potential effect on the development of phenotype of the advanced breeding lines of rice [17]. In present study Clifford and Stephenson (1975) criterion was considered which suggests that only first three canonical vectors or principle components play crucial role in depicting the patterns of genetic variations among breeding material, similar criterion was also adopted by Guei et al. (2005). Results of PCA revealed that first three components accounted for 61.85 per cent of total genetic variation, portraying intansic structure of variables being analysed. Similar trends in principal components were also observed by Vishnu et al. (2014), Ravi Kumar et al. (2015) and Gour et al. (2017) respectively for dissecting phenotypic variation with in breeding population and classifying genotypes based on potential morphological and quality traits. First principal component (PC-I) accounted for 26.75 % of total genetic variation present with in advanced breeding lines of rice. Traits such grain yield per plot and number of effective tillers per plant were reported to have maximum(0.45) contribution to genetic variation among all agro-morphological and quality traits under investigation followed by kernel breadth (0.35), total number of tillers per plant (0.26), grain yield per plant (0.22) and days to maturity (0.21) respectively. However plant height (-0.37) and panicle length (-0.23) were reported to contribute to total genetic variation negatively in PC-1. Principal component second (PC-II) was reported to account 19.49 % of total genetic variation. In PC-II, plant height (0.37) was reported to have maximum contribution genetic variation in second principal component followed by kernel breadth (0.35), days to 50 per cent flowering (0.32) and grain yield per plant (0.31) respectively [18]. However traits such as number of effective tillers per plant (-0.16), total number of plants per plant (0.17), kernel length (-0.41) and L/B ratio (-0.45) were reported to negatively add to total genetic variation in PCII respectively. Principal component third (PC-III) accounted for 15.60 % of total genetic variation available with in advanced breeding lines. In this component traits such as panicle length (0.55), kernel length (0.35), L/B ratio (0.33) and days to maturity (0.311) were reported to have maximum contribution toward total genetic variation as accounted by PC-III. However traits such as plant height (-0.15), total number of tillers per plant (-0.37) and number effective tillers per plant (-0.09) were reported to negatively contribute to total genetic variation accounted by PC-III. Fourth principal component was reported to account for 10.55 % of total genetic variation with in advanced breeding lines. In PC-IV traits which were reported to share maximum contribution to total genetic variation are days to 50 per cent flowering (0.26) followed by kernel breadth (0.17), grain yield per plot (0.16) and L/B ratio (0.014) respectively. Traits such as plant height (-0.01), number of effective tillers per plant (-0.23), panicle length (-0.63), kernel length (-0.29) and grain yield per plant (-0.44) respectively [19]. Principal component five (PC- V) was reported to account for 8.89 % of total genetic variation. In this component major contributors to total genetic variation were total number of tillers per plant (0.58) followed by days to maturity (0.45) and grain yield per plant (0.22) respectively, however traits such as panicle length (-0.04), 1000 grain weight (-0.29), kernel breadth (-0.20) and grain yield per plot (-0.38) were reported to negatively contribute to the genetic variation accounted by this component. Among six principal components or vetors first five components all together were reported to account for 81.30 % total genetic variation. Sathish and Senapati (2017) also reorted similar trends while investigating advanced breeding lines of rice grown in low land conditions of West Bengal. Mahendran et al. (2013) suggested that potential morphological and quality traits which remain side by side in various principal components and contributing maximum towards total available genetic variation with in breeding population have likelihood to remain together and provide ample scope and opportunity to be utilized in hybridization programmes [20].
| Agro-morphological and quality traits | PC-I | PC-II | PC-III | PC-IV | PC-V | PC-VI |
|---|---|---|---|---|---|---|
| Days to 50 percent flowering | 0.21 | 0.32 | 0.25 | 0.26 | 0.13 | 0.62 |
| PH (cm) | -0.37 | 0.37 | -0.15 | -0.01 | 0.18 | 0.28 |
| Total number of tillers per plant | 0.26 | -0.17 | -0.37 | 0.07 | 0.58 | 0.02 |
| Number of effective tillers per plant | 0.45 | -0.16 | -0.09 | -0.23 | 0.08 | 0.36 |
| DTM | 0.21 | 0.2 | 0.31 | 0.05 | 0.45 | -0.51 |
| PL(cm) | -0.23 | 0.16 | 0.55 | -0.29 | -0.04 | 0.04 |
| 1000 grain weight (g) | 0.15 | 0.1 | -0.29 | -0.63 | -0.29 | 0.005 |
| Kernel length (mm) | 0.03 | -0.41 | 0.35 | -0.29 | 0.18 | -0.04 |
| Kernel breadth (mm) | 0.35 | 0.35 | 0.02 | 0.17 | -0.2 | -0.13 |
| L/B ratio | 0.13 | -0.45 | 0.33 | 0.14 | -0.15 | 0.26 |
| Grain yield per plant (g) | 0.22 | 0.31 | 0.15 | -0.44 | 0.22 | 0.07 |
| Grain yield per plot (Kg) | 0.45 | 0.1 | 0.08 | 0.16 | -0.38 | -0.18 |
| Eigene value (Root) | 3.21 | 2.33 | 1.87 | 1.26 | 1.06 | 0.71 |
| Variance (%) | 26.75 | 19.49 | 15.6 | 10.55 | 8.89 | 5.93 |
| Cummulative variance (%) | 26.75 | 46.25 | 61.85 | 72.41 | 81.3 | 87.24 |
* PH: Plant height, DTM: Days to Maturity, PL:Panicle length
Table 6: Contribution of first six component axis to total genetic variation in advanced breeding lines.
From the foregoing discussion, it is apparent that there is a possibility for transferring the genes available within these advanced breeding lines of rice to create transgressive segregants with desired traits through selection and hybridization. Parents for hybridization should be chosen from separate clusters with a large inter and intracluster distance and the chosen parents should perform exceptionally well for the characters that contribute the most to genetic divergence. Mono-genotypic clusters with a single genotype could be used in hybridization programmes to exploit heterosis predominantly as testers. However, it is clear from the present study that crosses between genotypes from cluster II and those from cluster III, IV, V and VI are likely to express or demonstrate a high percentage of heterosis and generate potential recombinants with desirable traits. Further results of principal component or canonical vectors analysis classified the total genetic variation within advanced breeding lines by considering potential traits such as number of effective tillers per plant, grain yield per plot, plant height, panicle length and days to 50 per cent flowering, because these agro-morphological traits account for maximum contribution towards genetic diversity among advanced breeding lines. Selection for these traits will ensure efficient and goal oriented improvement of rice cultivers.
References
- Anonymous (2022) Digest of statistics. Directorate of economics and statistics, Govt of Jammu and Kashmir.
- Anonymous(2022) ICAR-National Rice Research Institute Cuttack, India.
- Clifford HT, Stephenson W (1975) An Introduction to Numerical Classification. Academic Press. London 229.
- Seetharaman RN, Sinha R, Banerjee SP (1988) Genetic divergence in rice. Indian J Genet 48:189-194.
- Dhakal K, Yadaw RB, Baral BR, Pokhrel KR, Rasaily S (2021) Agro-morphological and Genotypic diversity among rice germplasms under rainfed low land condition.J Agric Sci43: 466-478.
- Dubey K, Dwivedi DK, Singh A, Rampreet (2018) Genetic divergence studies among rice genotypes. J Pharmacogn Phytochem 2: 153-155.
- Gour L, Maurya SB, Koutu GK, Singh SK, Shukla SS, et al.(2017) Characterization of rice (Oryza sativa L.) genotypes using principal component analysis, including scree plot and rotated component matrix. Int J Chem Stud 5: 975-983.
- Guei RG, Sanni KA, Fawole AFJ (2005) Genetic diversity of rice (Oryza sativa). Agron Afr 5:17-28.
- Harman H H (1976) Modern factor analysis 3nd Ed. University of Chicago Press Chicago 376.
- Konate AK, Zongo A, Kam H, Sanni A, Audebert A (2016) Genetic variability and correlation analysis of rice (Oryza sativa L.) inbred lines based on agro-morphological traits.Afr J Plant Sci11: 3340-3346.
- Mahalanobis P C, (1928) On the need for standardisation in measurements on the living.Biometrika20: 1-31.
- Mahendran P, Veerabadhiran P, Robin S, Raveendran M (2015) Principal component analysis of rice germplasm.
- Mahendran R, Veerabadhiran P, Robin S, Raveendran M (2015) Principal component analysis of rice germplasm accessions under high temperature stress.Int J Agric Sci5: 355-359.
- Manohara KK, Singh NP (2013) Genetic divergence among rice landraces of Goa. ORYZA-An International Journal on Rice 5:100-104.
- Murthy BR, Arunachalam V (1996) The nature of genetic divergence in relation to breeding system in some crop plants. Indian Journal of Genetics 26:188-198.
- Rahman M, Acharya B, Sukla SN, Pande K (1997) Genetic divergence in low land rice genotypes. Oryza34: 209-212.
- Rao CR(1952) Advanced statistical methods in biometric research.
- Ravikumar BNVSR, NagaLumari P, Rao VRMP, Girija RPV, Satyanarayana N, et al. (2015) Principal component analysis and character association for yield components in rice (Oryza sativa L.) cultivars suitable for irrigated ecosystem. Current Biotica 9: 25-35.
- Sarwar G, Hossain MS, Harun-Ur-Rashid M, Parveen S(2015) Assessment of genetic variability for agro-morphological important traits in Aman rice (Oryza sativa L.).Int j appl sci biotechnol3:73-79.
- Vishnu VN, Robin S, Sudhakar D, Raveendran M, Rajeswari S, et al. (2014) Evaluation of Rice Genetic Diversity and Variability in a Population Panel by Principal Component Analysis. Indian J Sci Technol 7: 1555-1562.
Indexed at, Google Scholar, Crossref
Citation: Mushtaq D, Kumar B (2022) Multivariate Analysis among Advanced Breeding Lines of Rice (Oryza Sativa L.) Under Sub-Tropical Ecology of Jammu Region of J&K. J Plant Genet Breed 6: 134.
Copyright: © 2022 Mushtaq D, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Open Access Journals
Article Usage
- Total views: 2145
- [From(publication date): 0-2022 - Apr 02, 2025]
- Breakdown by view type
- HTML page views: 1780
- PDF downloads: 365