Multivariate Analyses of Phenotypic Diversity of Bread Wheat (Triticumaestivum L.) in the Highlands of Northern Ethiopia
Received: 07-Sep-2017 / Accepted Date: 19-Sep-2017 / Published Date: 27-Sep-2017 DOI: 10.4172/2329-8863.1000309
Abstract
An assessment of genetic variation within diverse germplasm is needed to allow more efficient genetic improvement. Forty-nine bread wheat genotypes were evaluated for 11 traits in simple lattice design at two locations to determine the extent of genetic diversity among the genotypes for grain yield and other agronomic traits. Mean squares of the traits studied showed statistically significant differences among the genotypes listed (P<0.01), indicating the presence of adequate variability. In the PC analysis, five PCAs explained 80.4% of total variability residing in the bread wheat genotypes. The first principal component, followed by the second, had the largest variance, and consequently explained much of the variability in the bread wheat genotypes. The traits which were important in these PCAs, plant height, grain yield, number of productive tillers, days to heading, spike length and number of spikelets per spike are the important traits in differentiating the genotypes. Average linkage cluster analysis classified the 49 genotypes into six clusters. Higher inter-cluster distance was exhibited between cluster I and III (D2=25.79**) followed by cluster II and IV (D2=22.82), and cluster II and III (D2=22.75), indicating wider genetic diversity among these clusters. Thus, future crossing program between members of cluster I with cluster III, and cluster II with III and IV could possibly result in heterosis in the F1, and a great deal of variability in the F2 generations.
Keywords: Bread wheat; Cluster analysis; Genetic distance; Grain yield; Principal component analysis
Introduction
Wheat, one of the globally important crops among the cereals, is cultivated approximately on 220 million and 9.9 million ha in the world and in Africa, respectively [1]. In Ethiopia, both bread and durum wheat species are produced by 4.6 million people on 1.66 million ha of land. It is the fourth important crop in area coverage next to tef, maize and sorghum; the third in productivity next to maize and rice [2].
The leading wheat producers in the world like Germany and France are capable of producing average yields of 7.4 and 7.2 t/ha, respectively [3], the world average of wheat production is about 3.25 ton per ha [1]. However, the average yield at production fields in Ethiopia is about 2.54 t/ha [2], which is much lower compared to the yield on experimental fields which is about 5 t/ha [4]. Thus, there is a need to further improve the yield of bread wheat.
Genetic diversity is indispensible for species adaptation to variable environmental conditions, it is valuable for germplasm collection and conservation and, the information also helps to study heterosis [5-7]. Plant breeding depends on the correct combination of specific alleles at the genetic loci present in a plants’ genome [8]. In order to access the alleles responsible for traits of agronomic importance, an assessment of genetic variation within diverse germplasm is needed to allow more efficient genetic improvement [9].
Therefore, this study was conducted with the objectives to study the extent of genetic diversity of bread wheat genotypes and to identify the important traits in distinguishing the genotypes.
Materials and Methods
Description of the study area
The experiment was conducted in 2015 at Jamma and Geregera research fields of Sirinka Agricultural Research Center. Jamma lies between 10°23' to 10°27' N latitudes and 39°7' to 39° 24'E longitudes, at an altitude of 2600 m.a.s.l. The dominant soil type is Vertisol and has PH of 6.0 with total rainfall of 720.5 mm. Geregera is located at an altitude of 2650 m.a.s.l, which lies between 38°45' E longitude and 11°46' N latitude with annual rainfall of 1105 mm. The soil type is characterized as Lithosol, and has pH of 5.6.
Planting materials
The planting materials include forty-nine bread wheat genotypes which are released varieties and elite materials obtained from Sinana and Kulumsa Agricultural Research Centers (Table 1).
| SN | Genotype | Maintainer | Genetic status | SN | Genotype | Maintainer | Genetic status |
|---|---|---|---|---|---|---|---|
| 1 | Alidoro | Holeta | Released | 26 | ETBW 8514 | Kulumsa | Elite line |
| 2 | Biqa | Kulumsa | Released | 27 | ETBW 8515 | Kulumsa | Elite line |
| 3 | Danda'a | Kulumsa | Released | 28 | ETBW 8516 | Kulumsa | Elite line |
| 4 | Digelu | Kulumsa | Released | 29 | ETBW 8517 | Kulumsa | Elite line |
| 5 | ETBW 6861 | Kulumsa | Elite line | 30 | ETBW 8518 | Kulumsa | Elite line |
| 6 | ETBW 6940 | Kulumsa | Elite line | 31 | ETBW 8519 | Kulumsa | Elite line |
| 7 | ETBW 7038 | Kulumsa | Elite line | 32 | Gassay | Adet | Released |
| 8 | ETBW 7058 | Kulumsa | Elite line | 33 | Hidasse | Kulumsa | Released |
| 9 | ETBW 7101 | Kulumsa | Elite line | 34 | Hoggana | Kulumsa | Released |
| 10 | ETBW 7120 | Kulumsa | Elite line | 35 | Honqolo | Kulumsa | Released |
| 11 | ETBW 7147 | Kulumsa | Elite line | 36 | Hulluka | Kulumsa | Released |
| 12 | ETBW 7194 | Kulumsa | Elite line | 37 | Jeferson | Kulumsa | Released |
| 13 | ETBW 7213 | Kulumsa | Elite line | 38 | Kakaba | Kulumsa | Released |
| 14 | ETBW 7364 | Kulumsa | Elite line | 39 | King Bird | Kulumsa | Registered |
| 15 | ETBW 7368 | Kulumsa | Elite line | 40 | Mada-Wolabu | Sinana | Released |
| 16 | ETBW 7871 | Kulumsa | Elite line | 41 | Mekelle-3 | Mekele | Released |
| 17 | ETBW 7872 | Kulumsa | Elite line | 42 | Mekelle-4 | Mekele | Released |
| 18 | ETBW 8506 | Kulumsa | Elite line | 43 | Ogolcho | Kulumsa | Released |
| 19 | ETBW 8507 | Kulumsa | Elite line | 44 | Pavon-76 | Kulumsa | Released |
| 20 | ETBW 8508 | Kulumsa | Elite line | 45 | Shorima | Kulumsa | Released |
| 21 | ETBW 8509 | Kulumsa | Elite line | 46 | Sofumar | Sinana | Released |
| 22 | ETBW 8510 | Kulumsa | Elite line | 47 | TAY | Adet | Released |
| 23 | ETBW 8511 | Kulumsa | Elite line | 48 | UTQUE96/3/PYN/BAU//MILLAN | Sinana | Released |
| 24 | ETBW 8512 | Kulumsa | Elite line | 49 | WORRAKATTA/PASTOR | Sinana | Released |
| 25 | ETBW 8513 | Kulumsa | Elite line |
Table 1: Description of the 49 bread wheat genotypes included in the diversity analysis.
Experimental design and trial management
The treatments were arranged in 7 × 7 simple lattice design replicated twice. The individual plot area was 1.2 m × 2.5 m (3 m2) with six rows for each genotype. The spacing between plots and rows were 0.4 m and 0.2 m, respectively. A 1.5 m space was left between blocks. Planting was done with seed rate of 150 kg/ha. Both Diammonium phosphate (DAP) and Urea fertilizers were applied at the rate of 100 kg/ha. DAP was applied at planting while 1/3 of the Urea was applied at planting and 2/3 at mid tillering stages.
Data collection and statistical analyses
Eleven quantitative traits were recorded. Ten plants were randomly chosen from the central four rows for recording data on plant height (cm), number of productive tillers per plant, spike length (cm), number of spikelets per spike and number of grains per spike. While the data for days to heading and maturity, thousand-seed weight (gm), biological yield (kg/m2), grain yield (qt/ha) and harvest index (%) were collected on plot basis considering the central four rows.
The data collected for each quantitative trait were subjected to analysis of variance (ANOVA) for simple lattice design using PROC LATTICE and PROC GLM procedures of SAS version 9.2 [10]. Homogeneity of error variances of each character were tested as per Hartley [11] before combining the data over the two locations. Principal component analysis was performed using correlation matrix by employing PAST 1.93 [12] to evaluate the contribution of each quantitative character in the total variation of genotypes. Factors (principal component axes) with eigenvalues values>1.0 were retained.
Mahalanobis [13] D2 statistics was used to estimate the genetic divergence in the bread wheat genotypes. The D2 statistics is defined by the following formula:
D2ij=(Xi-Xj) S-1 (Xi-Xj)
Where; D2ij=the squared-distance between any two genotypes i and j;
Xi and Xj=the vectors for the values for genotypes ith and jth genotypes;
S-1=the inverse of pooled variance covariance matrix within groups.
Testing the significance of the squared-distance values obtained for a pair of clusters was taken as the calculated value of x2 (chi-square) and tested against the tabulated x2 values at p-1 degree of freedom at 1% and 5% probability levels, where p=number of traits used for clustering genotypes.
The PROC CLUSTER of SAS 9.2, with average linkage method of clustering strategy, grouped and sorted the genotypes into clusters to form Dendrogam. Cubic clustering criterion (CCC), pseudo F and pseudo t2 statistics were used in determining the number of clusters.
Results and Discussion
Analysis of variance
In the study, the relative efficiency of RCBD was greater as compared to the simple lattice design for most characters, and sum of squares for blocks within replication were nonsignificant. As a result, analysis of variance was done using RCBD model. Homogeneity of error variance for all characters investigated was met. Consequently, ANOVA was run for the combined data over the two locations.
The location × genotype interaction was significant for all traits except number of spikelets per spike signifying differential response of the bread wheat genotypes across locations. Besides, significant (P<0.05) treatment differences were observed for all the characters studied, indicating the existence of genetic variability within the genotypes (Data not shown). The present investigation is in conformity with early findings in bread wheat [14-16].
Performance of the bread wheat genotypes
The success of a breeding program depends largely upon the amount of genetic variability present in the population and the extent to which the desired traits are heritable. Based on combined ANOVA over locations, the genotypes showed a wide range of variation for the studied traits (Table 2). Days to maturity ranged from 124 to 136 with mean value of 127.6 days, plant height ranged from 68-93.75 cm with mean value of 78.3 cm, number of grains per spike from 29.45-7 with mean value of 37.8 and thousand-seed weight from 34.8-48 g with mean value of 40.8 g. Also, grain yield ranged from 26.5-43.8 with mean value of 34.6 qt/ha. The sizable ranges of values for traits indicate good opportunity for bread wheat improvement. A similar wide range of variation among bread wheat genotypes in yield and yield related traits was reported by Kumar et al. [14] and Tesfaye et al. [15].
| Traits | Range | Mean ± SE | C.V (%) | LSD (5%) | R2 (%) |
|---|---|---|---|---|---|
| Days heading | 61-79.5 | 66.6±0.04 | 2.65 | 2.48 | 0.92 |
| Days maturity | 124-136 | 127.6±0.22 | 1.9 | 3.41 | 0.84 |
| Plant height (cm) | 68-93.75 | 78.3±0.084 | 6.45 | 7.09 | 0.84 |
| Number of productive tillers | 1.2-1.95 | 1.5±0.214 | 16.3 | 0.35 | 0.81 |
| Spike length (cm) | 6.4-10.9 | 7.5±0.104 | 8.97 | 0.94 | 0.86 |
| Number of spikelets per spike | 13 -17.4 | 14.6±0.78 | 7.3 | 1.48 | 0.84 |
| Number of grains per spike | 29-45.7 | 37.8±0.11 | 8.9 | 4.71 | 0.77 |
| Thousand-seed weight (gm) | 34.8-48 | 40.8±0.10 | 7.11 | 3.9 | 0.94 |
| Biological yield (kg/m2) | 1.8-2.70 | 2.12±0.123 | 10 | 0.299 | 0.89 |
| Harvest index | 0.26-0.36 | 0.31±0.16 | 9.66 | 0.043 | 0.94 |
| Grain yield (qt/h) | 26.5-43.8 | 34.6±0.20 | 13.7 | 6.66 | 0.95 |
Table 2: Estimates of range and means for 11 traits of 49 bread wheat genotypes, combined across the locations.
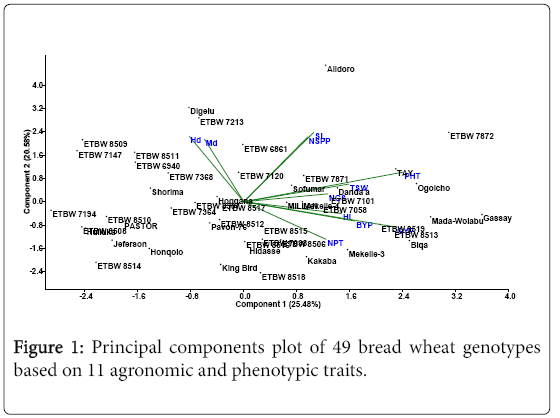
Principal components analysis
The main advantage of principal component analysis is extracting the most important information from the data table, compressing the size of the data set by keeping only this important information [17] and reducing the number of dimensions without much loss of information [18].
Out of the total 11 principal component axes (PCAs) extracted, five PCAs with eigenvalues greater than one were retained. The first principal component should have the largest possible variance, and therefore should explain the largest part of the variability [17]. According to the results, the first PCA explained 25.48% of the variation which is the highest of all the PCAs. The five PCAs all together explained 80.4% of total variability residing in the bread wheat genotypes, leaving the remaining 19.4% in the last six principal components (Table 3).
| Character | PCA1 | PCA2 | PCA3 | PCA 4 | PCA 5 |
|---|---|---|---|---|---|
| Days to heading | -0.165 | 0.451 | 0.434 | 0.212 | 0.117 |
| Days to maturity | -0.112 | 0.432 | 0.456 | 0.158 | 0.223 |
| Plant height | 0.483 | 0.204 | -0.184 | 0.02 | 0.157 |
| Number of productive tillers | 0.249 | -0.259 | 0.141 | 0.057 | 0.221 |
| Spike length | 0.212 | 0.479 | -0.245 | -0.096 | -0.332 |
| Number of spikelets per spike | 0.194 | 0.446 | -0.364 | -0.072 | -0.081 |
| Number of grains per spike | 0.255 | 0.055 | -0.147 | -0.096 | 0.811 |
| Thousand-seed weight | 0.319 | 0.122 | 0.357 | -0.058 | -0.201 |
| Biological yield | 0.337 | -0.133 | -0.023 | 0.7281 | -0.126 |
| Harvest index | 0.296 | -0.082 | 0.364 | -0.601 | -0.097 |
| Grain yield | 0.461 | -0.174 | 0.273 | 0.088 | -0.153 |
| Eigen value | 2.8 | 2.26 | 1.8 | 1.1 | 1.01 |
| Proportion (%) | 25.48 | 20.85 | 16.36 | 10 | 7.73 |
| Cumulative (%) | 25.48 | 45.22 | 63.04 | 73.49 | 80.4 |
Table 3: Vector loadings and percentage of explained variation by the first five PCAs. PCA=Principal component axis.
The factor loadings refer to the correlation between principal components and variables. A high correlation between PCAs and variables indicate that the variables are associated with the direction of the maximum amount of variation in the data set [17]. The data presented in Table 3 showed the most contributing characters are found in the first principal component which were plant height and grain yield; whereas number of productive tillers, days to heading, spike length and number of spikelets per spike contributed more in the second PCA. Likewise, days to maturity and thousand-seed weight in the third PCA; biological yield and harvest index in the fourth PCA; number of grains per spike in the fifth PCA, were the major contributing characters for variability to those principal components. Similar findings of grouping bread wheat genotypes by principal component analysis were reported [19-21].
It is also evidenced by the PCA plot (Figure 1) that genotypes depicted on the top part of the plot had high values of days to heading, spike length and spike number per spike; and those depicted on the right side of the plane had higher values of grain yield and plant height. The bread wheat genotypes at the top right side of the plane are expected to be late heading tall plants with higher number of spikelets and high yield. These genotypes could be used to develop superior varieties with desired traits. The fact that 80.4% of the variance is explained by five PCAs reveals the presence of wide variability in the genotypes for the character studied and it suggests ample opportunities for genetic improvement of bread wheat through direct selection, and utilization of the genotypes for future hybridization programs.
Genetic divergence and cluster analyses
Divergence analysis is performed using Mahalanobis [13] D2 distance to classify the diverse genotypes for hybridization purpose (Tables 4 and 5). The genetic improvement through hybridization and selection depends on the extent of genetic distance between parents. Chi-square test was used to declare significance of distance values using P-1 degrees of freedom where, P is the number of characters used in the study [22].
| Cluster | I | II | III | IV | V | VI |
|---|---|---|---|---|---|---|
| I | 10.29 | 25.79** | 7.36 | 13.31 | 16.07 | |
| II | 22.75* | 22.82* | 17 | 19.78* | ||
| III | 8.91 | 12.22 | 15.68 | |||
| IV | 7.06 | 17 | ||||
| V | 14.58 | |||||
| VI |
Table 4: Inter cluster D2 values among six clusters of 49 bread wheat genotypes using 11 quantitative traits. X2 =18.3, 23.2 (*, **) at 5% and 1% probability level, respectively.
| Cluster | Number of genotypes | Name of genotypes | Proportions (%) |
|---|---|---|---|
| I | 27 | Mekelle-3, Shorima, ETBW 7368, Hidasse, ETBW 8517, ETBW 7120, Sofumar, ETBW 7038, Kakaba, Pavon-76, ETBW 8512, ETBW 7871, Millan, ETBW 6861, ETBW 8506, Mekele-4, Biqa, ETBW 8513, ETBW 7058, ETBW 8519, ETBW 7101, ETBW 8515, Alidoro, Mada-Wolabu, Gassay, Ogolcho, Dandaa | 55.1 |
| II | 2 | TAY, ETBW 7872 | 4.08 |
| III | 15 | ETBW 8507, Jeferson, ETBW 8510, Honqolo, Hulluka, Pastor, ETBW 7194, ETBW 7364, ETBW 8507, ETBW 8514, ETBW 8509, ETBW 8516, King Bird, ETBW 8518, Hoggana | 30.6 |
| IV | 3 | ETBW 8511, ETBW 6940, ETBW 7213 | 6.12 |
| V | 1 | Digelu | 2.04 |
| VI | 1 | ETBW 7147 | 2.04 |
Table 5: Distribution and grouping of 49 bread wheat genotypes into different diversity classes based on D2 analysis.
Inter-cluster distance values (D2) between six clusters are presented in the (Table 4). The highest inter-cluster distance was exhibited between cluster I and III (D2=25.79**), followed by cluster II and IV (D2=22.82*), cluster II and III (D2=22.75*) and cluster II and VI (19.78*), indicating wider genetic divergence among the clusters. Thus, crossing of genotypes between members of cluster I with members of cluster III; members of cluster II with members of cluster III, IV and VI may produce high amount of heterotic expression in the F1’s and broad spectrum of variability in segregating (F2) populations. Genetic divergence in bread wheat genotypes reported by earlier workers [19-21, 23].
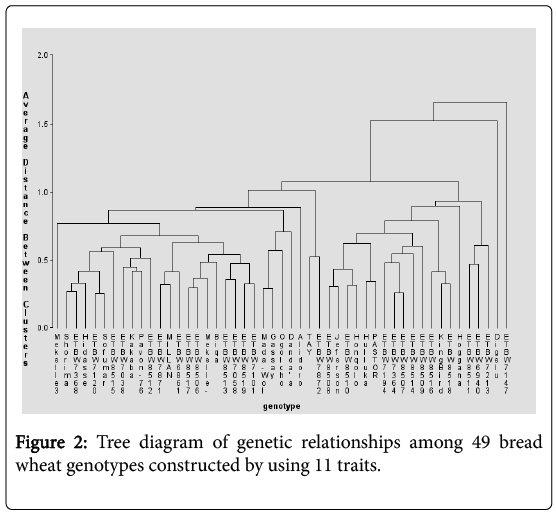
The dendrogram obtained from the cluster analysis through average linkage technique grouped the 49 genotypes into six clusters at about 47% similarity level based on D2 values computed using pooled mean trait data (Table 5 and Figure 2) which makes them moderately divergent. Related findings were reported by earlier workers [19-21]. Similarity between clusters is the average distance between all objects in one cluster and all objects in other cluster, whereby individuals within any cluster were more closely related than individuals in different clusters.
The genotypes were grouped in such a way that cluster I had the largest member of all clusters with 27 (55%) genotypes, followed by cluster III with 15 (30.6%) and cluster IV with 3 (6.04%) genotypes. In contrast, cluster V and cluster VI had the smallest member, constituting of one (2.04%) genotype each.
The mean values of the 11 quantitative traits for each cluster are presented in Table 6. Cluster I had a characteristic features of heading early, high grain yield per hectare and moderate high value in terms of harvest index. Cluster II had characteristic features of early maturity, tall plant type (plant height), high number of spike lets per spike, high grain yield, and relatively low harvest index as compared to other clusters. Cluster III showed earliness in days to heading and days to maturity as well as moderate grain yield. Cluster IV had characteristic features of relatively low values of biological yield and grain yield per hectare, and relatively moderate values in terms of characters studied. Cluster V had characteristic features of lateness in heading and maturity, high values in terms of number of grains per spike, thousand-seed weight and harvest index; on the other hand it had low value in terms of spike length and biological yield. Cluster VI had a characteristic feature of late heading and, short in plant height, high value in thousand-seed weight, and also characterized by high harvest index and grain yield per hectare.
| Cluster Means | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cluster | HD | MD | PH | NPTP | SL | NSPS | NGS | TSW | BY | HI | GY |
| I | 65.50 | 127.33 | 81.22 | 1.55 | 7.69 | 14.64 | 38.14 | 41.83 | 2.16 | 0.326 | 36.02 |
| II | 66.38 | 126.15 | 91.8 | 1.45 | 8.75 | 16.3 | 41.25 | 41.15 | 2.48 | 0.285 | 36.31 |
| III | 65.87 | 126.41 | 72.40 | 1.45 | 7.07 | 14.24 | 36.43 | 39.29 | 2.07 | 0.301 | 32.51 |
| IV | 71.33 | 132.67 | 75.73 | 1.48 | 8.07 | 14.93 | 39.60 | 38.50 | 2.00 | 0.300 | 30.96 |
| V | 79.50 | 136.30 | 80.9 | 1.45 | 6.80 | 15.00 | 45.70 | 43.10 | 1.78 | 0.330 | 31.75 |
| VI | 79.00 | 136.50 | 69.2 | 1.30 | 7.20 | 13.30 | 29.10 | 43.30 | 2.05 | 0.340 | 36.00 |
Table 6: Mean values of six clusters of 49 bread wheat genotypes for 11 characters. DH=days to heading, DM=days to maturity, PH=plant height, NPTP=number of productive tillers per plant, SL=spike length, NSPS=number of spikelets per spike, NGS=Number of grains per spike, BY=biological yield, HI=harvest index, TSW=thousand-seed weight, GY= grain yield.
As presented in Table 6, low and high mean values, respectively, were recorded between cluster VI and II for plant height, cluster VI and II for number of spikelets per spike, cluster VI and V for number of grains per spike, cluster IV and VI for thousand-seed weight, cluster V and II for biological yield, and for harvest index observed between cluster II and VI respectively. In addition to these the highest grain yield was obtained from cluster I, II and VI; however, comparatively low grain yield was obtained from cluster III, IV and V.
Summary and Conclusion
Overall variability within a crop is due to heritable and nonheritable components. The present study comprised 49 bread wheat genotypes that were evaluated at Jamma and Geregera environments to study genetic diversity in 49 bread wheat genotypes. The analysis of variance revealed significant differences (P
The principal component analysis revealed that five PCAs, with eigenvalues greater than unity, explained 80.4% of the total variability. The first principal component, followed by the second, had the largest variance, and consequently explained much of the variability in the bread wheat genotypes. The traits which were important in these PCAs, plant height, grain yield, number of productive tillers, days to heading, spike length and number of spikelets per spike are the important traits in differentiating the genotypes.
The highest inter-cluster distance was exhibited between cluster I and III (D2=25.79**), followed by cluster II and IV (D2=22.82) and cluster II and III (D2=22.75), indicating wider genetic diversity among the clusters. Therefore, initiating crossing program between members of cluster I with members of cluster III, and members of cluster II with members of cluster III and IV may produce a high amount of heterotic expression in the F1’s and broad spectrum of variability in segregating (F2) populations.
Acknowledgement
The authors would like to thank Sirinka Agricultural Research Center (SARC) for providing experimental fields. Thanks also due to Mr. Zerihun Tadesse for availing wheat seeds, to research assistants of SARC for the help on the research field.
References
- Central Statistical Agency (2015) Agricultural sample survey 2014/15: Report on area and production of major crops. Addis Ababa, Ethiopia.
- Yao J, Ma H, Yang X, Yoa G, Zhou M (2014) Inheritance of grain yield and its correlation with yield components in bread wheat (Triticumaestivum L.). African Journal of Biotechnology 13: 1379-1385.
- Ferede M, Worede F (2016) Grain yield stability and phenotypic correlation analysis of bread wheat (Triticumaestivum L.) genotypes in north western Ethiopia. Food Science and Quality Management 48: 51-59.
- Gao LZ (2003) The conservation of Chinese rice biodiversity: genetic erosion, ethnobotany and prospects. Genet. Resour Crop Evol 50: 17-32.
- Virk PS, Khush GS, Virmani SS (2003) Breeding strategies to enhance heterosis in rice. In: Virmani SS, Mao CX, Hardy B (eds.). Hybrid rice for food security, poverty alleviation and environmental protection. International Rice Research Institute. Proceedings of the 4th International Symposium on Hybrid Rice, Hanoi, Vietnam, Los Baños, pp: 21-29.
- Duran C, Appleby N, Edwards D, Batley J (2009) Molecular genetic markers: Discovery, applications, data storage and visualisation. Curr Bioinformatics 4: 16-27.
- Hoisington D, Khairallah M, Reeves T, Ribaut JM, Skovmand B (1999) Plant genetic resources: what can they contribute toward increased crop productivity. ProcNatlAcadSci USA 96: 5937-5943.
- Haussmann BIG, Parzies HK, Presterl T, Susic Z, Miedaner T (2004) Plant genetic resources in crop improvement. Plant Genetic Resources 2: 3-21.
- SAS Institute Inc (2008) SAS/STAT® 9.2 User’s Guide. SAS Institute Inc., Cary, NC, USA.
- Hartley HO (1950) The maximum F-ratio as a short cut test for heterogeneity of variances. Biometrika 37: 308-312.
- Hammer Ø, Harper DAT, Ryan PD (2001) Past: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4: 1-9.
- Mahalanobis PC (1936) On the generalized distance in statistics. Proc Nat InstSci 2: 49-55.
- Kumar N, Markar S, Kumar V (2014) Studies on heritability and genetic advance estimates in timely sown bread wheat (Triticumaestivum L.). Bioscience Discovery 5: 64-69.
- Tesfaye T, Genet T, Desalegn T (2014) Genetic variability, heritability and genetic diversity of bread wheat (Triticumaestivum L.) genotype in western Amhara region, Ethiopia. Wudpecker Journal of Agricultural Research 3: 26-34.
- Zeeshan M, Arshad W, Khan MI, Ali S, Tariq M (2014) Character association and casual effects of polygenic traits in spring wheat (Triticumaestivum L.) genotypes. International Journal of Agriculture, Forestry and Fisheries 2: 16-21.
- Zeeshan M, Arshad W, Khan MI, Ali S, Tariq M (2014) Character association and casual effects of polygenic traits in spring wheat (Triticumaestivum L.) genotypes. International Journal of Agriculture, Forestry and Fisheries 2: 16-21.
- Abdi H, Williams LJ (2010) Principal component analysis. WIREs Comp Stat 2: 433-459.
- Jolliffe IT (2002) Principal Component Analysis. 2nd ed. Springer Verlag, New York.
- Hailegiorgis D, Mesfin M, Genet T (2010) Genetic divergence analysis on some bread wheat genotypes grown in Ethiopia. J of Central European Agriculture 12: 344-352.
- Degewione A, Alamerew S (2013) Genetic diversity in bread wheat (Triticumaestivum L.) genotypes. Pak J Bio Sci 16: 1330-1335.
- Fellahi Z, Hannachi A, Bouzerzour H, Boutekrabt A (2013) Study of interrelationships among yield and yield related attributes by using various statistical methods in bread wheat (Triticumaestivum L.). International Journal of Agronomy and Plant Production 4: 1256-1266.
- Singh RK, Chaundry BD (1985) Biometrical methods in quantitative genetic analysis. Kalayani Publishers, Ludhiana, New Delhi, p: 318.
Citation: Getachew A, Worede F, Alamerew S (2017) Multivariate Analyses of Phenotypic Diversity of Bread Wheat (Triticum aestivum L.) in the Highlands of Northern Ethiopia. Adv Crop Sci Tech 5: 309. DOI: 10.4172/2329-8863.1000309
Copyright: © 2017 Getachew A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 7428
- [From(publication date): 0-2017 - Apr 02, 2025]
- Breakdown by view type
- HTML page views: 6410
- PDF downloads: 1018