Research Article Open Access
Multiplex LCMS Bioanalysis of Brentuximab Vedotin, Rituximab and Cetuximab towards Therapeutic Drug Monitoring Application by Combined Calibration Curve Using Fab-Selective Limited Proteolysis nSMOL
Noriko Iwamoto1, Megumi Takanashi1, Akinobu Hamada2 and Takashi Shimada1*1Technology Research Laboratory, SHIMADZU Corporation, Tokyo, Japan
2Division of Molecular Pharmacology & Pharmacokinetics, National Cancer Center Research Institute, Tokyo, Japan
- *Corresponding Author:
- Takashi Shimada
Technology Research Laboratory
SHIMADZU Corporation 1-3 Kanda-Nishiki-cho
Chiyoda-ku, Tokyo101-8448, Japan
Tel: 81335477630
Fax: 81335477636
E-mail: t-shima@shimadzu.co.jp
Received date: September 17, 2016; Accepted date: September 23, 2016; Published date: September 27, 2016
Citation: Iwamoto N, Takanashi M, Hamada A, Shimada T (2016) Multiplex LCMS Bioanalysis of Brentuximab Vedotin, Rituximab and Cetuximab towards Therapeutic Drug Monitoring Application by Combined Calibration Curve Using Fab-Selective Limited Proteolysis nSMOL. Clin Pharmacol Biopharm 5:164. doi:10.4172/2167-065X.1000164
Copyright: © 2016 Iwamoto N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Clinical Pharmacology & Biopharmaceutics
Abstract
Background: Recently, monoclonal antibody (mAb) bioanalysis using mass spectrometry has begun to be recognized as useful technology for mAbs measurement other than ELISA. We have recently exploited a high-precision method for bioanalysis of monoclonal antibody (mAb) using mass spectrometry. The method is nano-surface and molecular-orientation limited (nSMOL) proteolysis, which is useful for LCMS bioanalysis of many kinds of antibody drugs. Methods: nSMOL is Fab-selective limited proteolysis which consists of the difference of protease nanoparticle diameter (200 nm) and antibody resin pore diameter (100 nm). For limited proteolysis of antibody, Protein A resin (pore: 100 nm) slurry was added to plasma including monoclonal antibody, and the antibody Fc region was immobilized to the resin at 25°C for 10 min with gentle vortexing. Antibody-immobilized resin was washed with PBS, and limited proteolysis was performed with trypsin-conjugated FG beads (diameter: 200 nm). Limited proteolysis of Fab region on antibody was achieved by these two diameter difference. After nSMOL proteolysis, the generated peptides were collected by only simple filtration. Results: In this study, we have demonstrated that the first full validation dataset for bioanalysis using nSMOL of antibody-drug conjugate (ADC), Brentuximab vedotin, in human plasma using nSMOL proteolysis. Full validation using nSMOL proteolysis fulfilled criteria of guideline on bioanalytical method validation in pharmaceutical development for small molecule drug compounds. Conclusions: These results indicate that nSMOL is also significant method for precise quantification of ADC in plasma, such as Brentuximab vedotin. Furthermore, we report that nSMOL proteolysis is able to apply for not only single-analyte but also multi-analyte bioanalysis of each mAbs in plasma, so that, nSMOL proteolysis is feasible multiplex bioanalysis for many clinical pharmacokinetic study and therapeutic drug monitoring.
Keywords
Antibody drugs; Brentuximab vedotin; nSMOL; LCMS; Multiplex bioanalysis
Introduction
Monoclonal antibodies (mAb) are new innovation in various types of diseases treatment as a small molecule drugs. mAbs led from the effectiveness of the top sales of pharmaceutical products to dominant. However mAb has some issues, time consuming and expensive thing for manufacture, such as that hard to aim the intracellular targets for poor membrane permeability. On the other hand, conventional low molecular medicine has a low target selectivity, while there is no problem that these, there is a difficult problem to read, such as side effects and pharmacokinetics/pharmacodynamics (PK/PD). The antibody-drug conjugates (ADCs) are powerful tool to address this issue. Brentuximab vedotin is an ADC microtubule-disrupting agent (monomethyl auristatin E: MMAE) conjugate chimeric antibody against CD30, a protein on the surface of some Hodgkin lymphoma cells [1]. Brentuximab vedotin was currently approved by the US Food and Drug Administration (FDA) in 2011, and by the European Medicines Agency (EMA) in 2012 on the market based on its effectiveness in treatment-resistant Hodgkin’s lymphoma. The main mechanism by which Brentuximab vedotin exerts anti-tumor activity is due to the binding of the ADC to CD30-expressing cells, followed by internalization of the ADC-CD30 complex, and the release of MMAE via proteolytic cleavage, then MMAE disrupts the microtubule network, subsequently inducing cell cycle arrest and apoptosis of cancer cells [2,3]. Serum concentrations of Brentuximab vedotin by PK study were reported from 0.5 to 50 μg/mL [1,4].
Despite of high selectivity of mAbs, administration of mAbs carries the risk of immune-related adverse event such as acute anaphylaxis, infusion reaction, organ dysfunction, and the generation of anti-drug antibodies. Furthermore, the treatment of Trastuzumab for patients with metastatic gastric cancer, patients in the lowest concentrations of Trastuzumab had shorter overall survival than those in higher concentrations [5]. Thus, PK information is essential for individualized medicine and reduction of treatment costs.
For quantification of therapeutic mAbs in PK study, ligand binding assays such as enzyme-linked immunosorbent assay (ELISA) is the most widely use. However, quantification of ADCs by sandwich ELISA did not always allow for recovery of all ADCs. To address this issue, bioanalysis using LCMS is more appropriate for PK study with accurate information of ADCs concentration including biological sample. In the present study, we have developed nSMOL proteolysis for mAbs bioanalysis using LCMS technology [6]. nSMOL proteolysis is sensitive and selective analytical method for broadly applicable to regulated LCMS bioanalysis of mAb [7]. Full validation of five mAbs bioanalysis using nSMOL proteolysis had already reported. In this report, we evaluated that nSMOL proteolysis linked MS-based methodologies is available for quantification of ADCs with fulfilled all criteria of full validation guideline.
On the other hand, it is very difficult to develop an ELISA method capable of quantifying multiple proteins in one analysis. Multiple reaction monitoring (MRM) coupled with LCMS using a triple quadrupole mass spectrometer is a powerful method for quantitative measurement of specific proteins. MRM approaches can be multiplexed for many targets in one analysis. Such multiplexing analysis has been used in biomarker and proteome wide mechanism studies [8,9]. The multiplexing method is also highly valuable for not only research purpose but also development of biotherapeutics such as mAbs quantification and practical medical use in clinical core facility. Abundant protein depletion followed by limited fractionation and peptide level prior to LCMS analysis achieves limit of detection in lower concentration with high precision and accuracy. nSMOL proteolysis, pretreatment technology for mAbs bioanalysis, focused on protein limitation in final analyte, so that nSMOL proteolysis makes possible multiple mAbs bioanalysis in one assay.
Materials and Methods
Trypsin-immoblized glycidyl methacrylate (GMA)-coated nano-ferrite particle FG beads with surface activation by NHS group was purchased from Tamagawa Seiki (Nagano, Japan). Toyopearl AF-rProtein A HC-650F resin was from Tosoh (Tokyo, Japan). Brentuximab vedotin was obtained from Takeda Pharmaceutical (Osaka, Japan). Individual male and female human plasma EDTA-2K treated was from Kohjin Bio (Saitama, Japan). Trypsin gold was from Promega (Fitchburg, WI). n-octyl- β -D-thioglucopyranoside (OTG) was from Dojindo Laboratories (Kumamoto, Japan). P14R, internal standard synthetic peptide, was from Sigma Aldrich (St. Louis, MO). Ultrafree-MC GV centrifugal 0.22 μm filter was from Merck Millipore (Billerica, MA). Other reagents, buffers, and solvents were purchased from Sigma-Aldrich and Wako Pure Chemical Industries (Osaka, Japan).
Sequence confirmation of Brentuximab vedotin peptides by high-resolution MS analysis
Brentuximab vedotin (20 μg) was digested using trypsin (1 μg) in 150 μL of 25 mM Tris-HCl buffer (pH 8.0) at 37°C for 16 h. Trypsin reaction was quenched by adding 10% formic acid solution at a final concentration of 0.5%. For nSMOL reaction, 20 μg of Brentuximab vedotin was collected with 50 μL of PBS-substituted AF-rProtein A resin 50% slurry in 180 μL of PBS containing OTG with gentle vortexing at 25°C for 15 min. Protein A resin was collected on an Ultrafree filter (0.45 μm), washed at first twice using 300 μL of PBS containing 0.1% OTG, and then twice using 300 μL of PBS by centrifugation (10,000×g for 1 min), and finally substituted with 75 μL of 25 mM Tris-HCl (pH 8.0). nSMOL proteolysis was carried out using 1 μg trypsin on FG-beads with gentle vortexing at 37°C for 16 hrs. in saturated vapor atmosphere. After proteolysis, reaction was stopped by adding 10% formic acid at a final concentration of 0.5%. The peptide solution was easily collected by centrifugation (10,000×g for 1 min) to remove Protein A resin and trypsin FG-beads. These tryptic peptides from Brentuximab vedotin were analyzed using high-resolution liquid chromatography-linear ion trap time-of-flight MS (Nexera X2 ultra high performance liquid chromatograph and LCMS-IT-TOF, Shimadzu, Kyoto, Japan), and fragment ions were assigned using an in-house Mascot Server and Distiller (Matrix Science, London, UK) with Brentuximab vedotin amino acid sequence information. The LCMS conditions were as below: solvent A, 0.1% aqueous formic acid; solvent B, acetonitrile with 0.1% formic acid; column, L-column2 ODS, 2.1×150 mm, 2 μm, 10 nm pore (Chemicals Evaluation and Research Institute, Tokyo, Japan); column temperature, 40°C; flow rate, 0.2 mL/min; gradient program, 0-5 min: %B=3, 5-35 min: %B=3-30 gradient, 35-46 min: %B=95, 46-55 min: %B=3. MS and MS/MS spectra were obtained using desolvation line and heat block at 250 and 400°C, respectively. Nebulizer nitrogen gas flow was set to 3 liter/min. Drying gas pressure was 100 kPa. Ion accumulation time was 30ms for MS, and 70ms for MS/MS analysis. MS/MS analysis was performed using the automated data dependent mode. Ar pulse time into the ion trap cell was 125 μs. The electrode of collision-induced dissociation (CID) cell was set at -1.5 V.
Prediction of Brentuximab vedotin signature peptides
Amino acid sequences of mAb drugs were obtained from Kyoto Encyclopedia of Genes and Genomes (KEGG). Multiple alignment analysis was performed using the amino acid sequence of Brentuximab vedotin (KEGG DRUG entry D06409), Rituximab (D02994), Cetuximab (D03455), and Infiximab (D02598) by ClustalW algorithm on GENETYX software (GENETYX, Tokyo, Japan). By this in silico analysis, theoretical tryptic peptides containing the complementarity-determining region (CDR) sequence, amino acid substitution, positions of conserved cysteine residue, and insertion or deletion sequences were aligned.
Condition setting of multiple reaction monitoring (MRM) for Brentuximab vedotin peptides
The peptides were quantified using an LC-electrospray ionization- MS (LC-ESI-MS) with triple quadrupole (Nexera X2 and LCMS-8050, Shimadzu). The LCMS was operated as follows: solvent A, 0.1% aqueous formic acid; solvent B, acetonitrile with 0.1% formic acid; column, Shim-pack GISS C18, 2.1×50 mm, 1.9 μm, 20 nm pore (Shimadzu); column temperature, 50°C; flow rate, 0.4 mL/min; gradient program, 0-1 min: %B=1, 1-2 min: %B=1-23 gradient, 2-5 min: %B=23-35 gradient, 5-6 min: %B=95 with flow rate 1 mL/min, 5.8-6.2 min: %B=1 with flow rate 0.4 mL/min, and 6.2-7 min: %B=1. MS spectra were obtained with ESI probe temperature, desolvation line, and heat block at 350°C, 200°C, and 400°C, respectively. Nebulizer, heating, and drying nitrogen gas flows were set to 3, 15, and 5 L/min, respectively. The Dwell time was set to 10 ms for each transition. Information of MRM monitor ions of peptide fragments were from the measured values of structure-assigned fragments by high-resolution LCMS analysis. CID Ar partial pressure in the Q2 cell was set to 270 kPa. The electrode voltage of Q1 pre bias, collision cell Q2, Q3 pre bias, and parent and fragment ion m/z were performed optimization support software (LabSolutions, Shimadzu). For MRM transition, one fragment ion of b- or y-series was selected for quantitation, and two ions were selected for structural confirmation according to the optimized MRM ion yield (Table 1).
| Selected Peptide | Region | Optimal MRM condition | Role | |||
|---|---|---|---|---|---|---|
| Transition mass filter | Q1 [v] |
Collision [v] |
Q3 [v] |
|||
| VLIYAASNL ESGIPAR |
CDR of L-chain |
837.5→343.1 (y4+) | -26 | -21 | -24 | Quantitation |
| 837.5→213.1 (y2+) | -26 | -36 | -15 | Structure | ||
| 837.5→600.3 (y3+) | -26 | -32 | -22 | Structure | ||
| Selected peptide; peptide sequence for Brentuximab vedotin quantitation, Region; region of selected peptide, Transition mass filter; fragment ion m/z for quantitation from the parent ion m/z, Q1 [V]; voltage condition of the quadrupole cell Q1, Collision; electrode voltage of collision cell Q2, Q3 [V]; voltage condition of the quadrupole cell Q3, Role; purpose of each ion m/z. | ||||||
Table 1: MRM transition of Brentuximab vedotin signature peptide for bioanalysis
Valid sample preparation by nSMOL proteolysis
In the present study, we carried out a bioanalytical validation of Brentuximab vedotin in plasma using the nSMOL method as described in our previous report [6] with a part of improvement. Full validation of nSMOL proteolysis coupled with LC-MS/MS method was performed in accordance with the Guideline on Bioanalytical Method Validation in Pharmaceutical Development from Notification 0711-1 of the Evaluation and Licensing Division, Pharmaceutical and Food Safety Bureau, the Ministry of Health, Labour and Welfare, dated July 11, 2013. Briefly, validation sample sets were prepared and stored at - 20°C or -80°C for 24 h or longer before each validation assay. A 10 μL aliquot of Brentuximab vedotin-spiked human plasma was diluted 10-fold in PBS containing 0.1% OTG. The Ig fraction in plasma was collected with 25 μL of PBS-substituted AF-rProtein A resin (50% slurry) with gentle vortexing at 25°C for 15 min. Nonspecific absorption on Protein A resin was washed twice with 300 μL of PBS containing OTG and then with 300 μL of PBS, and then washed resin suspended in 75 μL of 25 mM Tris-HCl (pH 8.0) containing 10 fmol/μL P14R. nSMOL proteolysis was carried out using 10 μg trypsin on FG-beads with gentle vortexing at 50°C for 6 h in saturated vapor atmosphere. After nSMOL proteolysis, reaction was quenched by adding 10% formic acid at a final concentration of 0.5%. The peptide solution was recovered by centrifugation (10,000×g for 1 min) with to remove Protein A resin and trypsin FG-beads. These analytes were transferred into low protein binding polypropylene vials, and then performed LCMS analysis. The concentration of Brentuximab vedotin in plasma samples was set from 0.586 to 300 μg/mL with two-fold serially dilution for 10 calibration samples. The concentrations of LLOQ, low quality control (LQC), middle quality control (MQC), and high quality control (HQC) were 0.586, 1.76, 14.1 and 240 μg/mL, respectively.
Multiplex nSMOL assay preparation using combined calibration standards
For multiplex nSMOL assay development, we prepared the mixed standard set of Brentuximab vedotin, Rituximab, and Cetuximab in plasma from 0.586 to 300 μg/mL. And for the control subject, we also prepared the single standard of each three antibodies. The assay verification was carried out using LQC, MQC, and HQC sample of each antibody concentration sample. For MRM transition of Rituximab and Cetuximab, one fragment ion of b- or y-series was selected for quantitation, and two ions were selected for structural confirmation according to previously report [10,11].
Results
Structural confirmation of Brentuximab vedotin signature peptides by LCMS-IT-TOF MS and ClustalW analysis
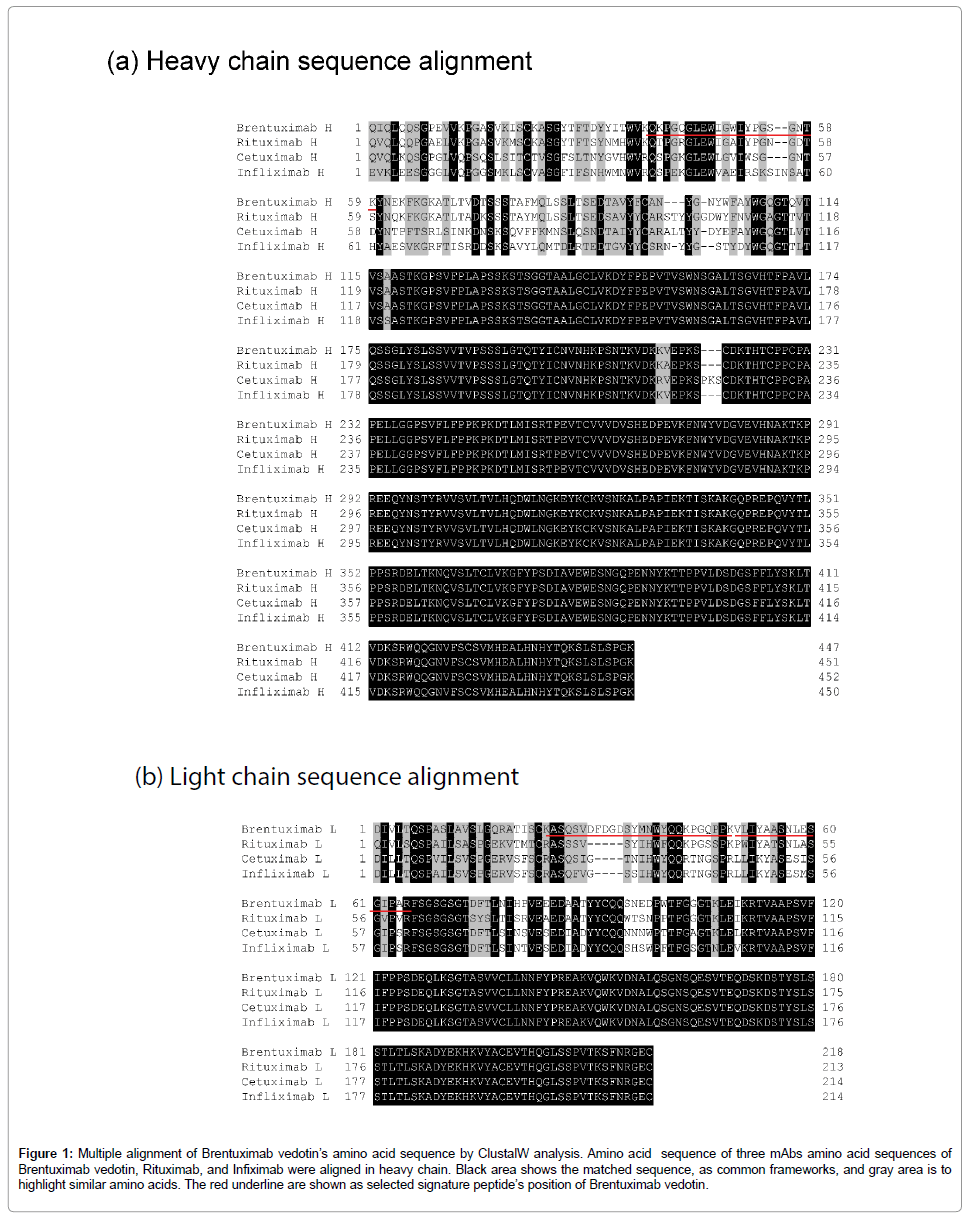
The identification of tryptic Brentuximab vedotin peptides by LCMS-IT-TOF MS and Mascot analysis showed that five tryptic peptides were identified using the nSMOL proteolysis, and three peptides were from CDR containing peptides and two peptides were from N-terminal containing peptides (Figures 1a and 1b). Finally, Brentuximab vedotin specific peptides were selected with some criteria for accurate quantitation like our present report. Finally, we have selected three candidate signature peptides, which is CDR containing peptides, VLIYAASNLESGIPAR, ASQSVDFDGDSYMNWYQQKPGQPPK, and QKPGQGLEWIGWIYPGSGNTK, for Brentuximab vedotin quantitation.
Figure 1: Multiple alignment of Brentuximab vedotin’s amino acid sequence by ClustalW analysis. Amino acid sequence of three mAbs amino acid sequences of Brentuximab vedotin, Rituximab, and Infiximab were aligned in heavy chain. Black area shows the matched sequence, as common frameworks, and gray area is to highlight similar amino acids. The red underline are shown as selected signature peptide’s position of Brentuximab vedotin.
Choice of the Brentuximab vedotin signature peptide in plasma for full validation
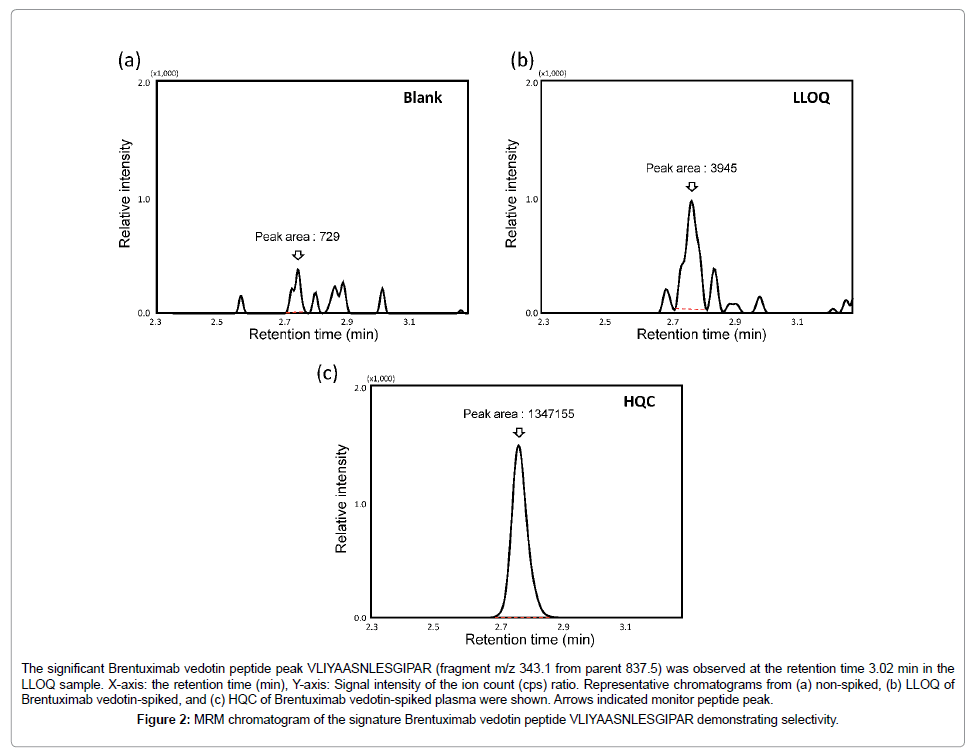
The analytical interference of candidate signature Brentuximab vedotin peptides in plasma matrix were analyzed by the nSMOL proteolysis coupled with LC-MS/MS. The peptide VLIYAASNLESGIPAR was selected as the signature peptide for Brentuximab vedotin quantification with no response in blank sample. We showed little interference from human plasma (Figure 2a) and a good correlation with Brentuximab vedotin concentrations. The optimized MRM transition of VLIYAASNLESGIPAR for quantitation and structure confirmation was shown in Table 1. Accordingly, following full validation, we selected VLIYAASNLESGIPAR peptide for quantitation of Brentuximab vedotin in plasma.
Full validation of Brentuximab vedotin
Selectivity and LLOQS: electivity is investigated using blank samples (without addition of Brentuximab vedotin and internal standard) obtained from at least 6 individual human plasma. Responses of these six individual human plasma controls from three males and three females were compared with the response of the LLOQ samples. Figure 2 showed representative chromatograms of the nonspiked, LLOQ, and HQC of Brentuximab vedotin monitor peptide. The chromatogram of P14R was shown in Figure S1. On the MRM chromatogram obtained from the blank sample on the same run, little interfering peak was observed at the retention time of Brentuximab vedotin (Figure 2). As a result, the LLOQ of Brentuximab vedotin was determined to be 0.586 μg/mL (Table 2). The response attributable to matrix components is less than 20% of the response at the LLOQ for the VLIYAASNLESGIPAR signature Brentuximab vedotin peptide and no response of the internal standard attributable to matrix is observed.
| Nominal concentration (µg/mL) | Back-calculated concentration (µg/mL) | Accuracy (%) | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | |
| 0.586 | 0.546 | 0.595 | 0.646 | 94.1 | 103 | 111 |
| 1.17 | 1.35 | 1.11 | 1.01 | 115 | 94.7 | 86.0 |
| 2.34 | 2.34 | 2.63 | 2.49 | 99.8 | 112 | 106 |
| 4.69 | 4.48 | 4.14 | 4.90 | 95.7 | 88.5 | 105 |
| 9.38 | 9.27 | 9.35 | 9.07 | 99.0 | 99.7 | 96.8 |
| 18.8 | 19.2 | 19.0 | 17.5 | 103 | 101 | 93.2 |
| 37.5 | 40.8 | 39.2 | 37.7 | 109 | 104 | 100 |
| 75.0 | 79.1 | 78.8 | 74.7 | 105 | 105 | 99.6 |
| 150 | 141 | 151 | 152 | 94.0 | 101 | 101 |
| 300 | 280 | 295 | 332 | 93.5 | 98.3 | 111 |
Table 2: Calibration curve
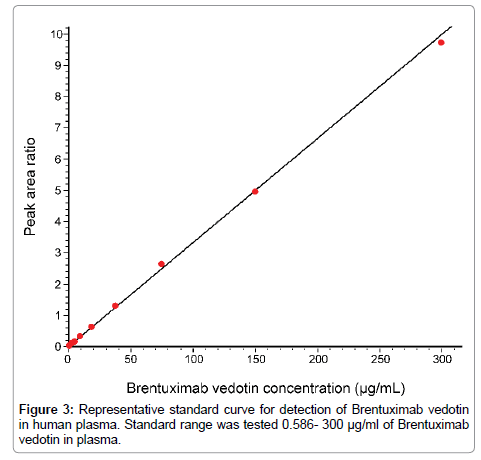
Linearity and calibration curve: Linearity of the bioanalysis using nSMOL proteolysis was demonstrated by analyzing ten calibration standards (zero sample, 0.586, 1.17, 2.34, 4.69, 9.38, 18.8, 37.5, 75.0, 150, and 300 μg/mL) using the linear regression model. The calibration plot of weighting was analyzed using the 1/area2 method. Linearity was confirmed with the accuracy of all the back calculated concentrations within the guideline criteria (LLOQ; ± 20% and other QC samples; ± 15%) and correlation coefficients greater than 0.99. The typical calibration curve of the Brentuximab vedotin peptide in plasma was shown in Figure 3. The calibration fit formulas of the triplicate runs were Y=0.0332X-0.00320 (r=0.998), Y = 0.0328X-0.00864 (r=0.999), Y = 0.0292X-0.00845 (r=0.994) (r: correlation coefficient). The accuracy at LLOQ was 94.1-111%, and other concentrations were 86.0-115% (Table 2).
Precision and accuracy in inter- and intra-assays: QC precision and accuracy were determined from analysis of human plasma validation sample at LLOQ, LQC, MQC, and HQC of Brentuximab vedotin as shown in Table 3. The intra-day and inter-day precision and accuracy were obtained by analyzing five replicates of QC samples at four concentration levels on three different days. As expected, precision and accuracy data were: run 1, 9.82% and 102% at LLOQ, 1.81-6.05% and 101-103% at other concentrations; run 2, 6.72% and 98.8% at LLOQ, 1.28-6.09%, 90.6-103% at other concentrations; run 3, 6.49% and 97.3% at LLOQ, 1.30%, 86.5-93.0% at other concentrations; intra-assay (N=15), 7.57% and 99.4% at LLOQ, 3.57-7.26%, 97.2-102% at other concentrations, respectively.
| Run | Nominal concentration | Concentration (µg/mL) | |||
|---|---|---|---|---|---|
| 0.586 | 1.76 | 14.1 | 240 | ||
| 1 | Observed | 0.626 | 1.94 | 14.2 | 243 |
| 0.564 | 1.71 | 14.7 | 247 | ||
| 0.512 | 1.86 | 14.3 | 259 | ||
| 0.629 | 1.70 | 14.4 | 240 | ||
| 0.657 | 1.72 | 14.0 | 242 | ||
| Mean | 0.598 | 1.79 | 14.3 | 246 | |
| SD | 0.06 | 0.11 | 0.26 | 7.60 | |
| CV (%) | 9.82 | 6.05 | 1.81 | 3.09 | |
| Accuracy (%) | 102 | 101 | 102 | 103 | |
| 2 | Observed | 0.577 | 1.78 | 12.6 | 242 |
| 0.514 | 1.81 | 12.5 | 212 | ||
| 0.606 | 1.80 | 13.0 | 217 | ||
| 0.611 | 1.84 | 12.9 | 211 | ||
| 0.588 | 1.79 | 12.7 | 211 | ||
| Mean | 0.579 | 1.80 | 12.7 | 219 | |
| SD | 0.04 | 0.02 | 0.21 | 13.32 | |
| CV (%) | 6.72 | 1.28 | 1.63 | 6.09 | |
| Accuracy (%) | 98.8 | 103 | 90.6 | 91.0 | |
| 3 | Observed | 0.623 | 1.85 | 14.0 | 256 |
| 0.560 | 1.77 | 14.2 | 250 | ||
| 0.520 | 1.75 | 13.7 | 248 | ||
| 0.571 | 1.75 | 13.9 | 249 | ||
| 0.578 | 1.81 | 14.0 | 245 | ||
| Mean | 0.570 | 1.79 | 13.9 | 250 | |
| SD | 0.04 | 0.04 | 0.18 | 4.04 | |
| CV (%) | 6.49 | 2.43 | 1.30 | 1.62 | |
| Accuracy (%) | 98.3 | 101 | 99.3 | 104 | |
| Mean (N=15) | 0.582 | 1.79 | 13.7 | 238 | |
| SD (N=15) | 0.04 | 0.06 | 0.73 | 17.28 | |
| CV (%) | 7.57 | 3.57 | 5.33 | 7.26 | |
| Accuracy (%) | 99.4 | 102 | 97.2 | 99.1 | |
Table 3: Precision and accuracy of VLIYAASNLESGIPAR
Matrix effect: The each plasma sample, six samples of males and females, was analyzed for matrix effect test at the LQC and HQC concentrations. The quantitative measure of matrix effect can be termed matrix factor (MF). MF was defined as the response ratio of the Brentuximab vedotin peptide in the presence of plasma prior to each sample preparation step was compared to in the absence of plasma. The average of matrix factors of P14R-corrected value at LQC and HQC was 3.14 and 0.90, respectively, which were within the accepted criteria of precision (CV) of<15% (Table 4).
| Analyte | Corresponding concentration (µg/mL) |
Blank matrix No. | P14R-normalized MF | Mean | SD | CV (%) |
|---|---|---|---|---|---|---|
| Brentuximab vedotin | 1.76 | M1 | 2.51 | 3.14 | 0.37 | 11.75 |
| M2 | 3.65 | |||||
| M3 | 3.23 | |||||
| F1 | 3.08 | |||||
| F2 | 3.15 | |||||
| F3 | 3.21 | |||||
| 240 | M1 | 0.965 | 0.897 | 0.08 | 8.74 | |
| M2 | 0.936 | |||||
| M3 | 0.775 | |||||
| F1 | 0.828 | |||||
| F2 | 0.914 | |||||
| F3 | 0.966 |
Table 4: Matrix effect
Carryover: The carryover test was performed by analyzing three replicates for Brentuximab vedotin signature peptide and evaluated by injecting blank samples after the highest concentration (300 μg/mL) sample. The carryover was calculated as percent response in the blank plasma compared with the LLOQ sample. In the nSMOL analysis, Brentuximab vedotin peptide carry over was 13.7-18.5% of the analyte response of Brentuximab vedotin peptide at the LLOQ and no analyte response of P14R was observed, confirming minimal levels of carryover influencing our analysis (Table 5).
| Compound | Run | Peak area | Peak area rate (%) | |
|---|---|---|---|---|
| LLOQ | Carry over sample |
|||
| Brentuximab vedotin | 1 | 2,656 | 363 | 13.7 |
| 2 | 3,945 | 729 | 18.5 | |
| 3 | 4,172 | 705 | 16.9 | |
| P14R | 1 | 155,665 | N.D. | N.D. |
Table 5: Carryover
Dilution integrity: The effect of dilution on the analysis of Brentuximab vedotin concentration using nSMOL proteolysis was assessed by using spiked validation human plasma at the concentration of 500 μg/mL. The 10- and 25-fold dilution of the five validation samples were analyzed within the calibration range. The precision and accuracy of the diluted samples were 4.71% and 94.5% 3.40% and 97.1%, for 10- and 25-fold dilutions, respectively. These values fulfilled the criteria of decision, indicating that absence of interference from dilution (Table 6).
| Nominal concentration (µg/mL) |
Dilution factor | Observed* (µg/mL) | Mean | SD | CV (%) |
Accuracy (%) |
|---|---|---|---|---|---|---|
| 500 | 10 | 51.1 | 472 | 2.23 | 4.71 | 94.5 |
| 46.6 | ||||||
| 45.5 | ||||||
| 46.4 | ||||||
| 46.6 | ||||||
| 500 | 25 | 20.1 | 485 | 0.66 | 3.40 | 97.1 |
| 19.7 | ||||||
| 18.9 | ||||||
| 18.6 | ||||||
| 19.9 |
Table 6: Dilution integrity
Stability: Stability of the analyte in matrix is evaluated at ambient conditions, five freeze-thaw cycles at -20°C and -80°C with at least 24 h of frozen time, storage stability short-term stability at room temperature for 4 h, long-term stability at -20°C and -80°C for 30 days prior to sample treatment, and post-preparative sample stability at 5°C for 24 and 48 h were demonstrated at the LQC and HQC concentrations. The mean accuracy in the measurements for freeze and thaw cycles stability at LQC and HQC in -20°C was 94.7% and 99.3%, respectively, and in -80°C was 100% and 102%, respectively. That for short-term stability (at room temperature for 4 h) at LQC and HQC was 89.8% and 88.5%, respectively. For long-term stability, the mean accuracy at LQC and HQC in -20°C was 97.0% and 101%, respectively, and in -80°C was 100% and 102%, respectively. Reinjection reproducibility after storage in autosampler is examined by quantification of LQC and HQC at 5°C for 24 h was 101% and 101%, respectively, and for 48 h was 104% and 102%, respectively (Table 7).
| Parameters for stability studies | Concentrations of Brentuximab vedotin in human plasma (µg/mL) | |||
| 1.76 | 240 | |||
| Mean (µg/mL) | Accuracy (%) | Mean (µg/mL) | Accuracy (%) | |
| Stability in plasma during freeze (-20°C) and thaw cycles | ||||
| Cycle 5 | 1.67 | 94.7 | 238 | 99.3 |
| Stability in plasma during freeze (-80°C) and thaw cycles | ||||
| Cycle 5 | 1.77 | 100 | 244 | 102 |
| Short-term stability in plasma for 4 h at room temperature | ||||
| 1.58 | 89.8 | 212 | 88.5 | |
| Long-term stability in plasma for 30 days at -20°C | ||||
| 1.71 | 97.0 | 242 | 101 | |
| Long-term stability in plasma for 30 days at -80°C | ||||
| 1.77 | 100 | 244 | 102 | |
| Processed sample stability in HPLC set at 5°C | ||||
| For 24 h For 48 h |
1.78 1.83 |
101 104 |
243 244 |
101 102 |
Table 7: Confirmation of QC sample for stability
Multiplex analysis for three mAbs in plasma: Three chimeric mAbs, Brentuximab vedotin, Rituximab, and Cetuximab, were mixed in plasma indicated concentrations. We performed nSMOL proteolysis against this three different mAbs containing plasma. For the LC-MS/MS method, signature peptides for quantification each mAb in a single run have already decided by previous our reports. These peptide sequences and the MRM transitions of three mAbs were showed in Table 8. The mixed ten calibration standards (zero sample, 0.586, 1.17, 2.34, 4.69, 9.38, 18.8, 37.5, 75.0, 150, and 300 μg/mL) of each mAb to calculate three QC levels. Linearity was observed with all the back-calculated concentration accuracy within acceptance criteria and correlation coefficients (r) being higher than 0.99. The QC samples were calculated using single analyte and multiple analyte standard curve. Under these conditions, QC accuracy and precision of this bioanalysis using nSMOL proteolysis fulfilled all guideline criteria, indicating that multiplex assay verification of mAbs bioanalysis using combined calibration standard was achieved by nSMOL proteolysis (Table 9).
| Analyte | Signature peptide | Optimal MRM condition | |||
|---|---|---|---|---|---|
| Transition mass [m/z] | Q1 [V] | Collision [V] | Q3 [V] | ||
| Brentuximab vedotin | VLIYAASNLESG IPAR | 837.5→343.1 | -26 | -21 | -24 |
| Cetuximab | SQVFFK | 378.2→540.3 | -17 | -15 | -28 |
| Rituximab | GLEWIGAIYPG NGDTSYNQK | 1092.1→1180.6 | -32 | -35 | -46 |
Table 8: Signature peptides from Brentuximab vedotin, Cetuximab, and Rituximab
| mAbs | Nominal concentration | Back-calculated concentration (µg/mL) | ||
|---|---|---|---|---|
| 1.76 | 14.1 | 240 | ||
| Brentuximab vedotin | Multiplex calculation1 | 1.92 | 15.4 | 253 |
| Multiplex calculation2 | 1.86 | 14.9 | 258 | |
| Multiplex calculation3 | 1.62 | 14.6 | 230 | |
| Single calculation1 | 1.76 | 15.2 | 264 | |
| Single calculation2 | 1.62 | 14.8 | 263 | |
| Single calculation3 | 1.64 | 13.4 | 252 | |
| Mean | 1.74 | 14.5 | 249 | |
| SD | 0.13 | 1.18 | 17.76 | |
| CV (%) | 7.60 | 8.15 | 7.12 | |
| Accuracy (%) | 99.2 | 103 | 104 | |
| Cetuximab | Multiplex calculation1 | 1.64 | 14.0 | 251 |
| Multiplex calculation2 | 1.53 | 12.4 | 263 | |
| Multiplex calculation3 | 1.92 | 15.0 | 244 | |
| Single calculation1 | 1.69 | 14.3 | 267 | |
| Single calculation2 | 1.62 | 13.0 | 268 | |
| Single calculation3 | 1.83 | 14.5 | 236 | |
| Mean | 1.71 | 13.9 | 255 | |
| SD | 0.14 | 0.98 | 14.07 | |
| CV (%) | 8.48 | 7.05 | 5.52 | |
| Accuracy (%) | 97.5 | 98.6 | 106 | |
| Rituximab | Multiplex calculation1 | 1.85 | 13.4 | 257 |
| Multiplex calculation2 | 1.68 | 14.7 | 261 | |
| Multiplex calculation3 | 1.82 | 12.7 | 225 | |
| Single calculation1 | 1.70 | 15.0 | 257 | |
| Single calculation2 | 1.69 | 12.4 | 259 | |
| Single calculation3 | 1.86 | 13.4 | 238 | |
| Mean | 1.77 | 13.6 | 249 | |
| SD | 0.15 | 1.09 | 16.45 | |
| CV (%) | 8.65 | 7.99 | 6.60 | |
| Accuracy (%) | 101 | 96.7 | 104 | |
Table 9: Precision and accuracy of mixed QCs containing Brentiximab vedotin, Cetuximab, and Rituximab
Discussion
In this report, we evaluated that nSMOL proteolysis linked MS-based methodologies is available for quantification of ADCs with fulfilled all criteria of full validation guideline. And we succeeded the individual antibody quantitation in plasma using combined multiplex nSMOL assays.
We previously developed a practical and versatile analytical method to assess mAb drugs concentration in plasma. There are abundant proteins, such as serum albumin and endogenous IgGs in plasma, so that pellet digestion and other previous reported methods have a problem of matrix interference and overall LCMS sustainability. In addition, for quantification of mAbs using LCMS is need of optimal signature peptide monitor. The nSMOL method is designed as solid-solid proteolysis for Fab-selective limited proteolysis. nSMOL proteolysis makes it possible to collect mAbs signature peptides including CDR domain and provide widely acceptable for mAbs bioanalysis on PK studies. Consequently, the nSMOL coupled with LC-MS/MS based bioanalysis assay has some advantages compared to ELISA and to the traditional method that the nSMOL proteolysis is introduced to improve the selectivity, robustness, and dynamic range, minimizing the analyte contamination, quickly method development, multiplex analysis for multiple analytes, and independent of biological taxonomy source.
ADCs are widely developed as next-generation dugs. Brentuximab vedotin is the first approved ADCs for treating Hodgkin lymphoma. Only Brentuximab was not promising enough to warrant clinical development but target molecule CD30, which is highly expressed in Hodgkin lymphoma, ALCL, cutaneous T-cell, and other selected lymphoid tumors, is highly potent anti-CD30 ADC for the treatment of CD30-positive malignancies. The anti-target molecule antibody is useful for development of ADCs as next-generation antibody, so that ADCs will become to apply in more diverse diseases. The ADCs bioanalysis using nSMOL helps the evolution of ADCs at preclinical and clinical PK study. As demonstrated by our present validation reports, nSMOL was able to provide a robust determination of ADC Brentuximab vedotin in human plasma. This is the first report describing a bioanalytical assay using nSMOL proteolysis for the quantification of ADC in human plasma. One nSMOL proteolytic signature peptide including CDR was significantly validated according to requirements of Guideline on Bioanalysis Method Validation in Pharmaceutical Development for small molecule LCMS. In addition, the analytical run of the mixed mAbs also demonstrated nSMOL proteolysis coupled with LCMS analysis robustness by the QC precision and accuracy.
Recently, mAb-based cancer treatments are diversifying the therapeutic applications of mAbs, indicating combined administration therapy of mAbs and other types of mAbs treatment after pretreated mAb [12,13]. Thus, the demand for multiplexing mAbs bioanalysis increases in both preclinical and clinical PK studies. Yet, ELISA could not measure multiple analyte in one analysis. The appropriate methodology for multiplex mAbs bioanalysis using LCMS has been demanded. We demonstrated that nSMOL proteolysis coupled with LCMS analysis has potential advantage of multiplex mAbs bioanalysis. mAbs bioanalysis using LCMS, unlike ELISA, it is possible to quantify multiple analyte in one analysis. It is demonstrated that the nSMOL proteolysis is suitable for multiplex mAbs bioanalysis using LCMS by the results of the validation of precision and accuracy.
On the other hand, biosimilars, such as Rituximab, Adalimumab, and Trastuzumab, have been developing, and Infliximab biosimilars had already approved in 2012. The evaluation of the comparability of biosimilars to the innovator drug should fulfill the guidelines laid out by the FDA and EMA. nSMOL proteolysis because of high selectivity and a short development time of the analytical method is convenient method for comparative PK studies of original and biosimilar production.
Monoclonal antibody-based therapy becomes highly divergent in cancer and autoimmune diseases. However, each mAb drugs may act differently on each patient that is used. It is important for individualized medicine to measure trough levels of the drug. For feasible and potentially optimal individual treatment, it is imperative to develop bioanalysis methods which make possible precise PK studies.
Acknowledgement
This work was partly supported by Grants-in-Aid from the Ministry of Health and Labor Sciences Research of Japan (AH).
References
- Younes A, Bartlett NL, Leonard JP, Kennedy DA, Lynch CM, et al. (2010) Brentuximab vedotin (SGN-35) for relapsed CD30-positive lymphomas. N Engl J Med 363: 1812-1821.
- Sutherland MS, Sanderson RJ, Gordon KA, Andreyka J, Cerveny CG, et al. (2006) Lysosomal trafficking and cysteine protease metabolism confer target-specific cytotoxicity by peptide-linked anti-CD30-auristatin conjugates. J Biol Chem 281: 10540-10547.
- Francisco JA, Cerveny CG, Meyer DL, Mixan BJ, Klussman K, et al. (2003) cAC10-vcMMAE, an anti-CD30-monomethyl auristatin E conjugate with potent and selective antitumor activity. Blood 102: 1458-1465.
- Ogura M, Tobinai K, Hatake K, Ishizawa K, Uike N, et al. (2014) Phase I / II study of brentuximabvedotin in Japanese patients with relapsed or refractory CD30-positive Hodgkin's lymphoma or systemic anaplastic large-cell lymphoma. Cancer Sci 105: 840-846.
- Yang J, Zhao H, Garnett C, Rahman A, Gobburu JV, et al. (2013) The combination of exposure-response and case-control analyses in regulatory decision making. J Clin Pharmacol 53: 160-166.
- Iwamoto N, Shimada T, Umino Y, Aoki C, Aoki Y, et al. (2014) Selective detection of complementarity-determining regions of monoclonal antibody by limiting protease access to the substrate: nano-surface and molecular-orientation limited proteolysis. Analyst 139: 576-580.
- Jenkins R, Duggan JX, Aubry AF, Zeng J, Lee JW, et al. (2015) Recommendations for validation of LC-MS/MS bioanalytical methods for protein biotherapeutics. AAPS J 17: 1-16.
- Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, et al. (2009) Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol 27: 633-641.
- Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. (2007) Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics : MCP 6: 2212-2229.
- Iwamoto N, Takanashi M, Umino Y, Aoki C, Hamada A, et al. (2016) Application of nano-surface and molecular-orientation limited proteolysis to LC-MS bioanalysis of cetuximab. Bioanalysis 8: 1009-1020.
- Iwamoto N, Takanashi M, Hamada A, Shimada T. (2016) Validated LC/MS Bioanalysis of Rituximab CDR Peptides Using Nano-surface and Molecular-Orientation Limited (nSMOL) Proteolysis. Biol Pharm Bull 39: 1187-1194.
- Younes A, Santoro A, Shipp M, Zinzani PL, Timmerman JM, et al. (2016) Nivolumab for classical Hodgkin's lymphoma after failure of both autologous stem-cell transplantation and brentuximabvedotin: a multicentre, multicohort, single-arm phase 2 trial. Lancet Oncol 17: 1283-1294.
- Armand P, Shipp MA, Ribrag V, Michot JM, Zinzani PL, et al. (2016) Programmed Death-1 Blockade With Pembrolizumab in Patients With Classical Hodgkin Lymphoma After BrentuximabVedotin Failure. J Clin Oncol.
Relevant Topics
- Applied Biopharmaceutics
- Biomarker Discovery
- Biopharmaceuticals Manufacturing and Industry
- Biopharmaceuticals Process Validation
- Biopharmaceutics and Drug Disposition
- Clinical Drug Trials
- Clinical Pharmacists
- Clinical Pharmacology
- Clinical Research Studies
- Clinical Trials Databases
- DMPK (Drug Metabolism and Pharmacokinetics)
- Medical Trails/ Drug Medical Trails
- Methods in Clinical Pharmacology
- Pharmacoeconomics
- Pharmacogenomics
- Pharmacokinetic-Pharmacodynamic (PK-PD) Modeling
- Precision Medicine
- Preclinical safety evaluation of biopharmaceuticals
- Psychopharmacology
Recommended Journals
Article Tools
Article Usage
- Total views: 12599
- [From(publication date):
November-2016 - Apr 02, 2025] - Breakdown by view type
- HTML page views : 11627
- PDF downloads : 972