Multilocus Sequence Typing and Molecular Detection of Phenol-soluble Modulin in Biofilm-positive Staphylococcus epidermidis Isolated from Paediatric Blood Culture
Received: 12-Feb-2019 / Accepted Date: 22-Feb-2019 / Published Date: 28-Feb-2019 DOI: 10.4172/2332-0877.1000396
Abstract
Aim: Staphylococcus epidermidis is a significant coagulase-negative staphylococci obtained from blood culture samples. However, there is limited information about phenol-soluble modulin (PSM), which is associated with virulence in S. epidermidis and its genetic relatedness in Nigeria. This study observed the presence of phenolsoluble modulin mec (psm-mec) gene and the multilocus sequence typing (MLST) of biofilm-positive Staphylococcus epidermidis (BPSE).
Method: Twenty-two biofilm-positive S. epidermidis isolates obtained from paediatric blood culture at three hospitals in north-west and north-central Nigeria were evaluated for the molecular detection of the psm-mec gene using conventional polymerase chain reaction (PCR). The biofilm formation was previously assessed by molecular detection of the intercellular adhesion (icaA) gene and the methicillin resistance using cefoxitin disk agar diffusion. Internal fragments of the respective seven housekeeping genes was sequenced for 21 BPSE strains and matched with the central MLST database.
Results: Out of 22 BPSE, only 4.5% had the psm-mec gene and it was methicillin resistant. About 91% methicillin resistance was observed among the psm-mec negative BPSE strains. Twenty-one BPSE strains were sequence type 1 (ST1) based on the MLST analysis of the seven housekeeping genes.
Conclusion: Considering the very low presence of psm-mec gene, the BPSE may be carrying the ß-type PSMs related to biofilm formation and dissemination, and not the cytolytic a-type PSMs common in the aggressive form of S. epidermidis. Phenol-soluble modulin methicillin resistance island-encoded peptide toxin is involved in sepsis related methicillin-resistant S. epidermidis. The strains are genetically related to each other.
Keywords: Phenol-soluble modulin; Methicillin resistance; Staphylococcus epidermidis ; Biofilm-positive; Multilocus sequence typing
Introduction
Almost all Staphylococcal strains secrete multifunctional peptide toxins known as phenol-soluble modulins (PSMs) with multiple studies ascribing their importance to the pathogenesis of staphylococci [1]. The secreted virulence factor located precisely in the methicillin resistance mobile genetic elements of certain Staphylococcal cassette chromosome (SCCmec) subtypes is known as phenol-soluble modulins [2]. Generally, members of the PSM family contribute to biofilm structuring and detachment, promote inflammatory, receptormediated responses in different human cell types and also exhibit strong cytolytic activity to neutrophils [3]. Phenol-soluble modulins are the main cytolytic and inflammatory toxins of S. aureus and S. epidermidis [4].
The common types are the alpha (α-type) and beta (β-type) PSMs. Alpha-phenol-soluble modulins (PSMα) are a novel class of miniature peptides having amphipatic alpha-helical structure produced by most staphylococci species [4]. The α-type PSMs have the cytolytic capacity and a shorter size having between 20-25 amino acids which contribute to the evasion of the innate immune system. They have properties that mimic strong surfactants and are majorly produced by Staphylococcal strains, in particular S. aureus and S. epidermidis [5,6]. The β-type PSMs are non-cytolytic, active in biofilm formation and have a size of approximately 40-45 amino acids.
The first discovery of PSMs in S. epidermidis was on the phenol layer following the hot aqueous phenol extraction of culture supernatant in the course of investigating factors associated with proinflammatory activity in a macrophage cell line [7]. The role of PSMs in staphylococci includes the activation and lyses of neutrophils to evade immune damage, support the production of proinflammatory cytokine, enhance biofilm structuring and detachment, facilitate biofilm-associated Staphylococcal infection and effectively eradicate competing bacteria [5,8].
The Psm-mec belongs to the amphipathic phenol-soluble modulin family and it is the only Staphylococcal toxin encoded by the psm-mec gene localized in the SCCmec element, which also contains the mecA genes, regulatory elements, recombinase genes, and some resistance genes [4,8].
Multilocus sequence typing (MLST) is a reference genotyping method that is suitable for analysing the evolution and population genetics of organisms [9,10]. It is a vital and investigatory tool in bacterial typing, and studying of staphylococci evolution, population dynamics, mode of bacterial dissemination and epidemiology [11]. A major advantage of MLST as a reference method is due to the unambiguous nature of DNA sequences stored easily on internetlinked databases with the corresponding clinical information of the isolate [12,13].
Genetic nucleotide sequence variations are identified and correctly interpreted in understanding the molecular epidemiology of S. epidermidis [14]. Previously, three different schemes for MLST proposed for S. epidermidis comprising varying genes were analysed but they proved difficult in studying the clinically diverse S. epidermidis strains globally and did not yield adequate resolution for understanding its epidemiology [15]. However, an improved MLST scheme was developed based on the comparative study of the previous MLST schemes reported and this was recognized and adopted as the MLST scheme for S. epidermidis [13].
The MLST scheme focuses on seven housekeeping genes present in all isolates of each bacterial species, their varying location in the bacterial chromosome and functions. Every distinct nucleotide sequence at a locus is designated an allele. A sequence type (ST) refers to each unique classification of alleles across the loci [9,16].
Materials and Methods
This study was carried out on 22 biofilm-positive Staphylococcus epidermidis isolated from children under five with bacteraemia attending the neonatal and paediatric units of University of Abuja Teaching Hospital, Federal Medical Center, Keffi and Murtala Mohammed Specialist Hospital, Kano between 2009 and 2016. The initial study involved the detection of methicillin resistance using cefoxitin disk agar diffusion method, biofilm formation by Christensen biofilm assay and the molecular detection of the intercellular adhesion (icaA) gene in the S. epidermidis isolates obtained from positive paediatric blood cultures of seven hospitals in northern Nigeria [17].
Ethical approval was obtained from the Research Ethics Committee of the hospitals and consent was given by the International Foundation against Infectious disease in Nigeria (IFAIN) to gain access to the S. epidermidis isolates obtained from positive blood culture in the respective hospitals.
Polymerase chain reaction (PCR) amplification of the psm-mec gene was performed using primers designed for this study. The 25 μl PCR reaction consisted of 12.5 μl of Midas Mix with Taq DNA polymerase (Monserate Biotechnology Group), 1 μl diluted cell culture, 9.5 μl of DNase/RNase-free distilled water and 1 μl each of psm-mec forward primer (5’-TGCATATGGATTTCACTGGTGTTA-3’) and reverse primer (5’-CGTTGAATATTTCCTCTGTTTTTTAGTTG-3’). The psm-mec gene used had a product size of 157 bp and the PCR amplification was run at 560C annealing temperature for the 22 biofilm-positive S. epidermidis strains. One of the strains was lost [17].
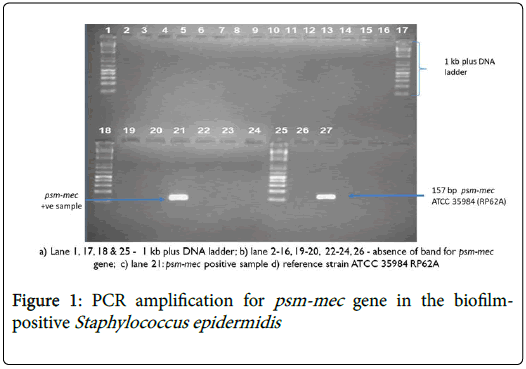
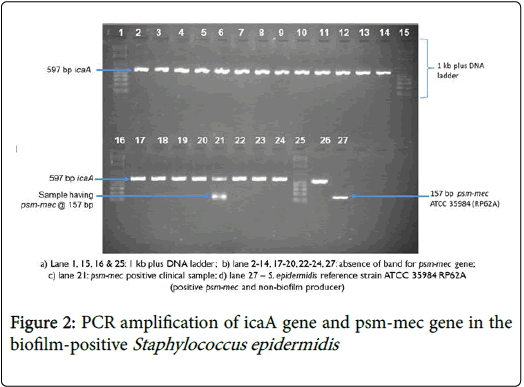
Gel concentration was increased to 2% agarose gel by measuring 2 g of agarose powder into 100 ml of tris-borate-EDTA (TBE) due to the small product size of psm-mec gene used (Figure 1). The S. epidermidis RP62A (ATCC 35984) was used as a positive control. A second duplex PCR run for psm-mec gene was done with icaA primers [17] at 560C to ensure that each of the PCR products had the respective icaA-positive S. epidermidis (Figure 2).
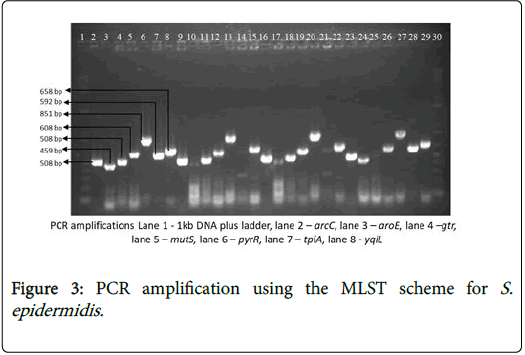
The internal fragments of the seven housekeeping genes (arcC, aroE, gtr, mutS, pyrR, tpi, and yqiL ) were amplified by PCR, using the specific primers with varying amplicon size ad annealing temperature (Table 1) with the purified PCR products as the DNA template [18].
| Gene and function | Primer | Sequences (5'-3') | Amplicon Size (bp) | Annealing T°C |
|---|---|---|---|---|
| Carbamate Kinase (arcC) | arcC-F | TGTGATGAGCACGCTACCGTTAG | 508 | 58°C |
| arcC-R | TCCAAGTAAACCCATCGGTCTG | |||
| Shikimate dehydrogenase (aroE) | aroE-F | CATTGGATTACCTCTTTGTTCAGC | 459 | 60°C |
| aroE-R | CAAGCGAAATCTGTTGGGG | |||
| ABC transporter (gtr) | gtr-F | CAGCCAATTCTTTTATGACTTTT | 508 | 60°C |
| gtr-R | GTGATTAAAGGTATTGATTTGAAT | |||
| DNA mismatch repair protein (mutS) | mutS-F3 | GATATAAGAATAAGGGTTGTGAA | 608 | 60°C |
| muts-R3 | GTAATCGTCTCAGTTATCATGTT | |||
| Pyrimidine operon regulatory protein (pyrR) | pyrR-F2 | GTTACTAATACTTTTGCTGTGTTT | 851 | 60°C |
| pyrR-R4 | GTAGAATGTAAAGAGACTAAAATGAA | |||
| Triosephosphate isomerase (tpiA) | tpi-F2 | ATCAATTAGACGCTTTAGTAAC | 592 | 58°C |
| tpi-R2 | TTAATGATGCGCCACCTACA | |||
| Acetyl coenzyme A acetyltransferase (yqiL) | yqiL-F2 | CACGCATAGTATTAGCTGAAG | 658 | 60°C |
| yqiL-R2 | CTAATGCCTTCATCTTGAGAAATAA |
Table 1: Seven housekeeping genes for multi locus sequence typing of S. epidermis.
A collection of 21 biofilm producing S. epidermidis isolates were analysed by MLST protocol. The PCR was performed with 25 μl reaction volume, composed of 1 μl each of the forward and reverse primer, 12.5 μl of Midas mix and 9.5 of RNAse/DNase free sterile water.
The conditions for running the PCR amplification involved an initial denaturation of 95°C for 3 min; 30 cycles of 95 °C for 30 s, annealing of primers between 58-60°C for 1 min based on the seven housekeeping genes, extension at 72 °C for 1 min; and a final extension of 72°C for 10 min.
The PCR products were analysed for DNA band separation by running 1% agarose gel electrophoresis in tris-borate-EDTA buffer (Figure 3). The amplified PCR products were analysed by 0.8% agarose gel electrophoresis with ethidium bromide. It was visualized and captured on UV transilluminator (Bio Rad Gel Doc). The size of the amplicon was determined by comparing with the GeneRuler™ 1 kb plus DNA ladder [19,20].
The amplified fragments were purified using DNA Clean and ConcentratorTM-5 Kit (Zymo Reasearch). DNA quantification was done using the nanodrop spectrophotometer to measure the purified PCR products in nanogram/microliter. The DNA sequencing was performed using Illumina HiSeq 2500 sequence analyser at the Genomics Core Facility of University of Nebraska Medical Centre, Omaha, Nebraska. The sequence analysis was done using NCBIBLAST tool to analyse the seven housekeeping genes and confirm their specificity to the corresponding GenBank sequence check for correct amplification of the primers [10].
The nucleotide sequencing gives confirmation and precise identification of most of the bacteria, and it was run on the basic local alignment search tool (BLAST) against S. epidermidis genome to find the regions of sequence similarity. The sequence types (ST) of the 21 BPSE strains were assigned based on the MLST analysis of allelic profiles of the seven housekeeping genes by sequence comparison using the available S. epidermidis MLST database.
Results
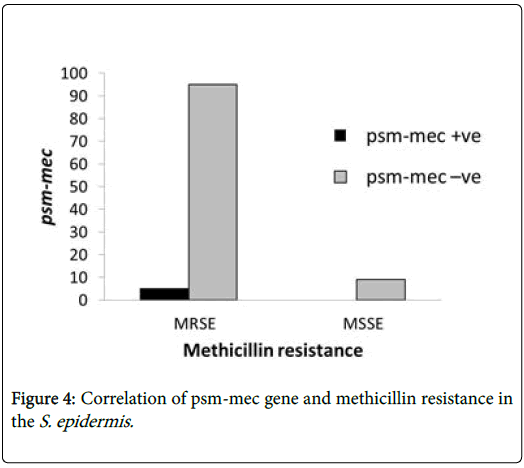
Out of 22 biofilm-positives S. epidermidis , only 4.5% carried the virulence determinant psm-mec gene while 95.5% did not have the psm-mec gene (Table 2). The S. epidermidis strain having the psm-mec gene was also methicillin resistant (Figure 3). About 91% of the psmmec negative S. epidermidis isolates were methicillin resistant (Figure 4).
|
Parameter |
Value |
|---|---|
|
psm-mec+ve |
4.5% (1) |
|
psm-mec-ve |
95.5%(21) |
Table 2: Detection of psm-mec gene in biofilm-positive Staphylococcus epidermidis.
The allelic profile match of the 21 biofilm-positive S. epidermidis isolates analysed using MLST were 1, 2, 2, 2, 1, 1, 10 resulting in sequence type 1 (ST1). The strain designation was DEN19 originating from Denmark. A detailed result of the biofilm-producing S. epidermidis strains in this study is indicated in (Table 3).
| Number | Isolate ID | (Grace et al., 2018) | (This study) | ||
|---|---|---|---|---|---|
| MRSE | icaA@597bp | psm-mec @157bp | Sequence Type (ST) | ||
| 1 | R1 | + | + | - | ST1 |
| 2 | R11 | + | + | - | ST1 |
| 3 | R14 | + | + | - | ST1 |
| 4 | R21 | + | + | - | ST1 |
| 5 | R28 | + | + | - | ST1 |
| 6 | R44 | + | + | ** | ** |
| 7 | R46 | + | + | - | ST1 |
| 8 | R56 | + | + | - | ST1 |
| 9 | R75 | + | + | - | ST1 |
| 10 | R79 | + | + | - | ** |
| 11 | R87 | + | + | - | ST1 |
| 12 | R100 | + | + | - | ST1 |
| 13 | R120 | - | + | - | ST1 |
| 14 | R191 | + | + | - | ST1 |
| 15 | R201 | + | + | - | ST1 |
| 16 | R219 | + | + | - | ST1 |
| 17 | R231 | + | + | - | ST1 |
| 18 | R235 | + | + | - | ST1 |
| 19 | R240 | + | + | - | ST1 |
| 20 | R245 | + | + | - | ST1 |
| 21 | P13 | + | + | - | ST1 |
| 22 | P32 | + | + | - | ST1 |
| 23 | P71 | + | + | - | ST1 |
Table 3: Detailed breakdown of MRSE, icaA, psm-mec and MLST of the S. epidermis strains.
Discussion
Staphylococcus epidermidis is known to produce phenol soluble modulins (PSM) that exhibit selective antimicrobial activity against other organisms and are proinflammatory cytolysins. This selective activity enables them to dominate other microbes as a skin flora [18]. Only 4.5% of the twenty-two biofilm-positive S. epidermidis had the psm-mec gene in this study. It is believed that the β-type PSMs produced by S. epidermidis participate mainly in biofilm formation and facilitates in vivo dissemination of biofilm-associated infection, and in vitro biofilm detachment. However, this process can be blocked by anti-PSM β antibodies [1,21].
The production of cytolytic α-type PSMs is very low in S. epidermidis and it is a significant cause of its lower pathogenicity and aggressiveness when compared to S. aureus [22]. The PSM methicillin resistance island-encoded peptide toxin is involved in sepsis related methicillin-resistant S. epidermidis [23].
Presence of virulence determinants in S. epidermidis is very limited but the persistence of clinical infection is often associated with genes that code for methicillin resistance and biofilm formation [24,25]. This agrees with the study by Qin, et al., (2017) [23] indicating that S. epidermidis has a high resistance rate of methicillin resistance.
Using the multilocus sequence typing (MLST), a collection of 21 biofilm-producing S. epidermidis isolates analysed in the study were sequence type 1 (ST1). This suggests a low level of genetic diversity and the strains are genetically related to each other. ST1 has also been reported in Denmark, Iceland, Canada and the United States of America. However, in a study by Sharma, et al., (2014) [26], the most commonly identified sequence types (ST) among the S. epidermidis obtained from blood cultures were ST 2 (21%) and ST 5 (14%).
Although the S. epidermidis strains used in this entire study for MLST were biofilm producers, a study carried out in Finland on the MLST of methicillin-resistant S. epidermidis obtained from bacteraemic patients were all sequence type ST2, indicating a genetic relatedness [25]. The S. epidermidis sequence type 2 (ST2) is characterized by antibiotic resistance, biofilm formation and a very flexible genetic background. It seems to be the common nosocomial clone obtained from neonates [11].
The use of MLST is becoming necessary in understanding global epidemiology of most pathogenic bacteria [9]. The central MLST database for S. epidermidis is progressively developing and contains distinct catalogue of genetic variants, which aids the direct identification of nucleotide sequence variation. However, there is still limited knowledge in the population genetics in Nigeria.
Conclusion
Considering the very low presence of psm-mec gene, the biofilmpositive S. epidermidis strains may be carrying the β-typ PSMs related to biofilm formation and dissemination, and not the cytolytic α-type PSMs common in the aggressive form of S. epidermidis . Additionally, S. epidermidis does not produce aggressive virulence factors or toxins; and the cytolytic PSMs when present are not secreted at a significantl toxic level. The strains analysed are genetically related to each other.
References
- Berlon NR, Qi R, Sharma-Kuinkel BK, Joo HS, Park LP, et al. (2015) Clinical MRSA isolates from skin and soft tissue infections show increased in vitro production of phenol soluble modulins. J Infect 71: 447-457.
- Joo HS, Otto M (2014) The isolation and analysis of phenol-soluble modulins of Staphylococcus epidermidis. Methods mol biol 1106: 93-100.
- Qin L, McCausland JW, Cheung GY, Otto M (2016) Psm-mec: A virulence determinant that connects transcriptional regulation, virulence and antibiotic resistance in Staphylococci. Front Microbiol 7: 1293.
- Monecke S, Engelmann I, Archambault M, Coleman DC, Coombs GW, et al. (2012) Distribution of SCCmec-associated phenol-soluble modulin in staphylococci. Mol Cell Probes 26: 99-103.
- Li S, Huang H, Rao X, Chen W, Wang Z, et al. (2014) Phenol-soluble modulins: Novel virulence-associated peptides of staphylococci. Future Microbiol 9: 203-216.
- Cheung GUC, Duong AC, Otto M. (2012) Direct and synergistic hemolysis caused by Staphylococcus phenol-soluble modulins: Implications for diagnosis and pathogenesis. Microbes Infect 14: 380-386.
- Mehlin C, Headley CM, Klebanoff SJ (1999) An inflammatory polypeptide complex from Staphylococcus epidermidis: Isolation and characterization. J Exp Med 189: 907-918.
- Chatterjee SS, Chen L, Joo HS, Cheung GY, Kreiswirth BN, et al. (2011) Distribution and regulation of the mobile genetic element-encoded phenol-soluble modulin Psm-mec in methicillin-resistant Staphylococcus aureus. PLoS ONE 6: e28781.
- Inouye M, Conway TC, Zobel J, Holt KE (2012) Short read sequence typing (SRST): Multilocus sequence types from short reads. BMC Genomics 13:338.
- Larsen MV, Cosentino S, Rasmussen S,  Friis C, Hasman H et al. (2012) Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50: 1355-1361.
- Dong Y, Speer CP, Glaser K (2018) Beyond sepsis: Staphylococcus epidermidis is an underestimated but significant contributor to neonatal morbidity. Virulence 9:621-633.
- Hiramatsu K, Katayama Y, Matsuo M, Sasaki T, Morimoto Y, et al. (2014) Multi-drug-resistant Staphylococcus aureus and future chemotherapy. J Infect Chemother 20: 593-601.
- Miragaia M, Carrico J A, Thomas JC, Couto I, Enright MC, et al. (2008) Comparison of molecular typing methods for characterisation of Staphylococcus epidermidis: Proposal for clone definition. J Clin Microbiol 46: 118-129.
- Lin T, Lin L, Zhang F (2014) Review of molecular typing methods of pathogens. Open J Med Microbiol 4: 147-152.
- Thomas JC, Vargas MR, Miragaia M, Peacock SJ, Archer GL, et al. (2007) Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J Clin Microbiol 45: 616-619.
- Belen A, Pavon I, Maiden MCJ (2009) Multilocus sequence typing. Methods Mol Biol 551: 129-140
- Grace JA, Olayinka BO, Onaolapo JA, Hassan-Hanga F, Munir H, et al. (2018) Methicillin resistance and biofilm formation of Staphylococcus epidermidis in blood culture isolates from children under five: A multicenter study in Nigeria. Clin Microbiol 7: 6.
- Fey PD (2014) Staphylococcus epidermidis: Methods and Protocols, New York: Humana Press.
- Thomas JC, Robinson DA (2014) Multilocus sequence typing of Staphylococcus epidermidis. In PD Fey, Staphylococcus Epidermidis: Methods and Protocols 61-68, New York: Humana Press, Springer.
- Hosseinzadeh S, Dastmalchi H (2014) Staphylococcal species associated with bovine mastitis in the North West of Iran: Emerging of caogulase-negative staphylococci. Int J Vet Sci Med 2: 27-34.
- Otto M (2014) Staphylococcus epidermidis pathogenesis. In Fey PD Staphylococcus epidermidis: Methods and Protocols 17-26, London:Â Humana Press.
- Da F, Joo HS, Cheung GC, Villaruz AE, Rohde H, et al. (2017) Phenol-soluble modulin toxins of Staphylococcus haemolyticus. Front Cell Infect Microbiol 7: 206.
- Qin L, Da F, Fisher EL, Tan DC, Nguyen TH, et al. (2017) Toxin mediates sepsis causedbymethicillin-resistant Staphylococcus epidermidis. PLoS Pathog 13: e1006153.
- Conlan S, Mijares LA, NISC Comparative Sequencing Program, Becker J, Blakesley RW, et al. (2012) Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol 13: R64.
- Ibrahem S, Salmenlinna S, Lyytikainen O, Vaara M, Vuopio-Varkila J (2008) Molecular characterisation of methicillin-resistant Staphylococcus epidermidis strains from bacteraemic patients Clin Microbiol Infect 14: 1020-1027.
- Sharma P, Satorius AE, Raff MR, Rivera A, Newton DW, et al. (2014) Multilocus sequence typing for interpretingbloodisolatesof Staphylococcus epidermidis. Interdiscip Perspect Infect Dis 2014: 1-4.
Citation: John-Ugwuanya GA, Obaro SK, Olayinka BO, Onaolapo JA, Hassan-Hanga F, et al. (2019) Multilocus Sequence Typing and Molecular Detection of Phenol-soluble Modulin in Biofilm-positive Staphylococcus epidermidis Isolated from Paediatric Blood Culture. J Infect Dis Ther 7: 396. DOI: 10.4172/2332-0877.1000396
Copyright: © 2019 John-Ugwuanya GA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 2944
- [From(publication date): 0-2019 - Feb 22, 2025]
- Breakdown by view type
- HTML page views: 2318
- PDF downloads: 626