MRI Hippocampal Subfield Volume Analysis: Comparison between Alzheimer’s Disease, Mild Cognitive Impairment, and Normal Aging Subjects in an Amyloid PET Project
Received: 01-Jan-2019 / Accepted Date: 24-Jan-2019 / Published Date: 31-Jan-2019 DOI: 10.4172/2161-0460.1000459
Abstract
Background: One of the most reliable biomarkers of Alzheimer’s Disease (AD) is hippocampal atrophy demonstrated by MRI. Based on neuropathological data showing a differential vulnerability of hippocampal subfields to AD processes, hippocampal subfield analysis could improve the diagnostic accuracy of AD or Mild Cognitive Impairment (MCI). The objective of this study was to demonstrate the difference of hippocampal subfield volumes by using automated analysis for discriminating between AD, MCI and Healthy Controls (HC).
Material and Methods: Fifty-three age-matched subjects including 15 AD (mean 72.3 ± 6.9 years), 9 MCI (mean 68.4 ± 3.9 years) and 29 HC (mean 69.5 ± 5.1 years) were recruited and underwent MRI (3-Tesla) with highresolution 3D-T1W. Quantitative volumetric analysis of hippocampal subfields was performed by using Freesurfer (v. 6.0) for comparing AD, MCI and HC subjects.
Results: There was a widespread pattern of subfield atrophy. Right subiculum, right presubiculum, bilateral molecular layer, and bilateral fimbria volumes were higher diagnostic efficacy than whole hippocampal volume for discriminating AD subjects from HC or MCI subjects. Only left fimbria volume was significantly reduced in MCI subjects compared to HC (normalized volume 47.7, sensitivity=66.7%, specificity=89.7%, AUC 0.709). The accuracy of left fimbria as the predictor of positive amyloid PET MCI was 75%.
Conclusion: The structural brain imaging using quantitative volumetric analysis of hippocampal subfield could be used for differentiating AD, MCI and HC. These findings may be helpful for early detection and follow up of AD and MCI patients in the clinical setting.
Keywords: Alzheimer’s disease; Mild cognitive impairment; Hippocampal subfield volume; Amyloid PET
Introduction
Alzheimer’s Disease (AD) is responsible for 60%–80% of the dementia patients [1]. Due to high individual cost, it was causing an enormous global socio-economic impact. In recent years, the researchers have strived to discover the non-invasive biomarkers that can reliably monitor disease progression and predict disease conversion in the future.
There are many modalities used for evaluation AD such as molecular imaging (FDG PET, amyloid PET) and advanced MR imaging including functional MRI; diffusion tensor imaging; and perfusion study [2-4]. However, these modalities need available and complex advanced processing methods for analysis. Structural brain imaging by using volume analysis either by visual evaluation or quantitative analysis by post-processing software is still helpful for evaluating AD patients and have the potential of reproducibility and easy to use in clinical practice in most healthcare.
Hippocampal atrophy has generally been found in AD [5,6] and also has been demonstrated in Mild Cognitive Impairment (MCI) with conversion to AD [7-9]. Hippocampal atrophy evidenced by MRI is one of the most validated biomarkers for AD. Based on neuropathological data showing a differential vulnerability of hippocampal subfields to AD processes, hippocampal subfield analysis should improve the diagnostic accuracy of AD and AD-converted MCI (cMCI).
Recently, many studies using varying techniques have attempted to measure the regional hippocampal degeneration by indirect surface-based approaches [10,11] and subfield volumetry from manual to automated methods [12]. The objective of this study was to demonstrate the differences of hippocampal subfield volumes by using automated subfield volumetric analysis in discriminating between AD, MCI and healthy control HC.
Materials and Methods
Study data, inclusion and diagnostic criteria
The study was approved by the institutional Review Board (Si 137/2015 and Si 059/2015, protocol number 651/2557(EC1)). Retrospective review data was performed, which derived from patients participating in Neurology and Geriatrics clinic in a tertiary hospital. A total of 53 subjects were included in this study (AD=15, MCI=9, HC=29). The demographics of the cohorts were shown in (Table 1).
Operational definition
AD
(1) Diagnosis established by National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association (NINCDS-ADRDA) for probable AD, (2) TMSE score less than 26, (3) Clinical Dementia Rating (CDR) score equal to or more than 0.5. Subjects were excluded from the study if any psychiatric or neurological illness other than AD was present.
MCI
(1) Subjects had TMSE scores between 24 and 30, (2) subjective memory complaint reported by the patient, family member or clinician with preserved activities of daily living, (3) CDR score of 0.5, (4) absence of dementia by NINCDS-ADRDA criteria.
HC
(1) TMSE scores between 24 and 30, (2) CDR score of 0, (3) no neurological or psychiatric illness, non-demented and normal ADLs.
MRI Acquisition
Standardized MRI data acquisition techniques were used for the study group on the 3T MR system (Ingenia, Philips Medical System, Best, the Netherlands) with a 32-channel head coil. The imaging protocol included a 3D high-resolution T1 weighted FFE covering the whole brain (field-of-view FOV 230×230×172 mm3, matrix size 352x352, voxel size 0.72×0.72×0.65 mm3, echo time TE /repetition time TR 4.8/9.8 ms; flip angle 8°, scan time 6 min).
Hippocampal subfield segmentation
Image analysis was accomplished by using the Freesurfer image analysis pipeline (version 6.0) a combination of ex vivo and in vivo MRI data [13]. Automated segmentation of the hippocampus was performed to define anatomical subfield labeling by using a Bayesian modeling approach and a computational model of the areas surrounding the hippocampus. An atlas mesh had previously been built and validated from manual delineations in ultra-high resolution MRI scans of 13 individuals. These delineations include the fimbria, presubiculum, subiculum, parasubiculum, CA1, CA2/3, CA4, molecular layer HP, GC-ML-DG (granule cell layer and molecular layer of dentate gyrus), HATA (hippocampus-amygdala-transition-area), hippocampal tail subfields and whole hippocampus as well as the hippocampal fissure.
All subfield measurements were normalized by the subject’s intracranial volume derived from Freesurfer using the following formula: volume_norm=volume_raw x 1,000/ICV in cm [3,14,15].
[18F] AV45 PET/CT Protocol
The detail of our in-house [18F] AV45 production and detailed chemical properties were as in the previous report [16]. The acquisition and image reconstruction protocols specific to our PET/CT scanner (Discovery STE, GE Healthcare, Wisconsin, USA), were as stated in ADNI GO [17] and ADNI 2 [18].Two nuclear medicine physicians with no access to clinical information, rated each amyloid PET image in consensus to perform binary interpretation of each data set as amyloid positive (increased retention of tracer in the cortical gray matter as shown either by the apparent loss of gray-white contrast within any two cortical regions, or intense uptake in at least one cortical region) and amyloid negative (preserved gray-white matter contrast with low retention of tracer in cortical gray matter), followed published criteria [19].
Statistical Analysis
Statistical analysis was carried out using PASW Statistics (Version 18. 0; SPSS inc., USA). Categorical variables were analyzed using the Chi-square test, and continuous variables were tested using ANOVA with Bonferroni post hoc comparisons. A p-value<0.05 was predetermined as statistically significant.
To assess the predictive performance of individual volumes, the Receiver Operating Characteristic (ROC) curve methodology was performed. The Area Under the Curve (AUC), sensitivity and specificity were calculated.
Results
Demographics, neuropsychological, and global clinical measurements
Fifty three subjects were included into the study (AD=15, MCI=9, HC=29). Amongst the subjects, clinical scores of the AD group were poorest, and of the HC group were best on TMSE, and CDR as expected (Table 1). The means TMSE of each group was 22.4 (SD 3.7) for AD, 26.8 (SD 1.8) for MCI, and 27.9 (SD 1.8) for HC.
| AD (n=15) | MCI (n=9) | HC (n=29) | p-value | |
|---|---|---|---|---|
| Gender (men/women) | 43684 | 7/2a | 44075 | 0.045 |
| Age (mean+SD) | 72.3 ± 6.9 | 68.4 ± 3.9 | 69.5 ± 5.1 | 0.187 |
| Education | 0.288 | |||
| - Primary school | 6 | 2 | 15 | |
| - Secondary school and High school | 4 | 1 | 3 | |
| - Higher than high school | 5 | 6 | 11 | |
| TMSE | 22.4 ± 3.7a,b | 26.8 ± 1.8 | 27.9 ± 1.8 | <.001 |
| ADAS-COG | 17.4 ± 10.6a,b | 8.2 ± 4.8a | 5.3 ± 2.6 | <.001 |
| CDR | <.001 | |||
| 0 | 0a | 0a | 27 | |
| 0.5 | 8a | 9a | 2 | |
| 1 | 7a,b | 0 | 0 | |
| ADCS-ADL | 60.4 ± 11.3a,b | 71.8 ± 3.9 | 73.7 ± 4.2 | <.001 |
| NPI (Min,Max) | 1.07(0,5) | 0.44(0,2) | 0.66(0,11) | 0.665 |
a Significant compared to HC
b Significant compared to MCI
TMSE: Thai Mental State Examination; ADAS-COG: Alzheimer’s Disease Assessment Scale-Cognitive Subscale
CDR: Clinical Dementia Rating Scale; ADL: Alzheimer’s Disease Cooperative Study Activities of Daily Living (ADCS-ADL); NPI: Neuropsychiatric inventory
Table 1: Demographic, clinical and neuropsychological data in AD, MCI and HC subjects.
Hippocampal subfields
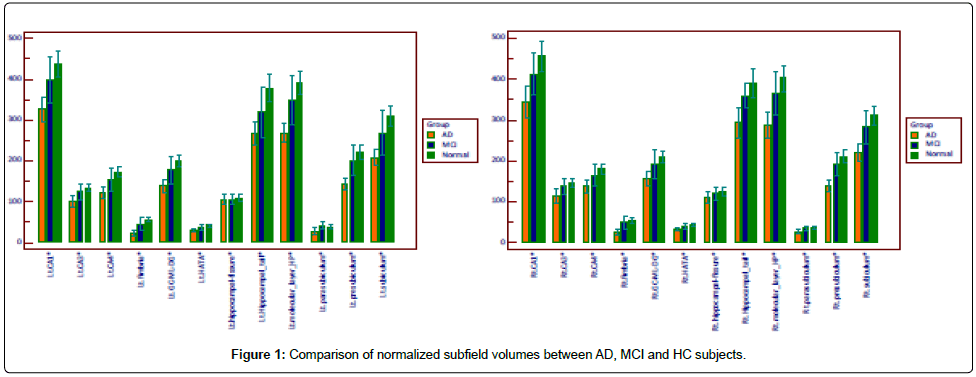
All of the subfield volumes, except for hippocampal fissure, were significantly different between AD group and MCI or HC group (p<0.5) (Table 2, Figure 1).
| AD (n=15) | MCI (n=9) | HC (n=29) | p value | |
|---|---|---|---|---|
| Lt. Hippocampal_tail | 270.4 ± 48.6a | 322.6 ± 69.4 | 379.5 ± 65.4 | <0.01 |
| Rt. Hippocampal_tail | 292.5 ± 62.1a | 355.9 ± 34.4 | 387.9 ± 71.2 | <0.01 |
| Lt. subiculum | 214.8 ± 38.8a+b | 270.5 ± 60.8 | 308.4 ± 47.2 | <0.01 |
| Rt. subiculum | 225.2 ±39.9a+b | 285.7 ± 47.0 | 313.3 ± 45.9 | <0.01 |
| Lt. CA1 | 329.3 ± 50.8a+b | 400.0 ± 61.6 | 437.7 ± 71.2 | <0.01 |
| Rt. CA1 | 347.3 ± 65.0a | 417.8 ± 59.8 | 456.3 ± 72.3 | <0.01 |
| Lt. hippocampal fissure | 106.8 ± 23.6 | 108.1 ± 15.5 | 107.1 ± 21.3 | 0.989 |
| Rt. hippocampalfissure | 111.2 ± 24.8 | 121.9 ± 16.4 | 123.8 ± 19.9 | 0.167 |
| Lt. presubiculum | 149.3 ± 31.3a+b | 201.7 ± 42.2 | 217.8 ± 34.9 | <0.01 |
| Rt. presubiculum | 142.2 ± 25.1a+b | 189.2 ± 32.3 | 210.0 ± 35.3 | <0.01 |
| Lt. parasubiculum | 30.3 ± 15.2 | 41.7 ± 11.0 | 38.3 ± 9.7 | 0.043 |
| Rt. parasubiculum | 28.5 ± 8.8a | 35.5 ± 6.9 | 37.3 ± 7.5 | 0.003 |
| Lt. molecular_layer_HP | 273.0 ± 43.3a+b | 350.7 ± 66.1 | 390.8 ± 56.4 | <0.01 |
| Rt. molecular_layer_HP | 289.2 ± 51.9a+b | 370.1 ± 61.2 | 404.0 ± 60.2 | <0.01 |
| Lt. GC-ML-DG | 141.4 ± 26.7a+b | 179.3 ± 37.3 | 199.3 ± 27.9 | <0.01 |
| Rt. GC-ML-DG | 156.4 ± 31.3a+b | 195.5 ± 39.5 | 210.7 ± 29.4 | <0.01 |
| Lt. CA2/3 | 102.4 ± 25.5a+b | 125.7 ± 21.4 | 133.6 ± 20.4 | <0.01 |
| Rt. CA2/3 | 113.0 ± 27.6a+b | 141.4 ± 24.9 | 146.8 ± 22.8 | <0.01 |
| Lt. CA4 | 124.1 ± 22.7a+b | 155.8 ± 29.6 | 171.7 ± 24.0 | <0.01 |
| Rt. CA4 | 136.8 ± 27.8a+b | 168.2 ± 30.7 | 180.0 ± 24.7 | <0.01 |
| Lt. fimbria | 24.2 ± 12.3a+b | 45.3 ± 17.4a | 57.8 ± 9.6 | <0.01 |
| Rt. fimbria | 28.8 ± 13.2a+b | 48.3 ± 17.8 | 56.6 ± 12.6 | <0.01 |
| Lt. HATA | 31.2 ± 7.6a | 37.8 ± 8.6 | 42.4 ± 7.8 | <0.01 |
| Rt. HATA | 34.1 ± 6.6a | 40.1 ± 7.0 | 45.3 ± 7.6 | <0.01 |
| Lt. Whole_hippocampus | 1690.2 ± 258.0a+b | 2131.2± 373.1 | 2377.2 ± 331.9 | <0.01 |
| Rt. Whole_hippocampus | 1793.9 ± 308.3a+b | 2247.6 ± 324.2 | 2448.4 ± 345.2 | <0.01 |
Normalized hippocampal subfields volume (absolute hippocampal subfield/intracranial volume) were used with Bon-ferroni pairwise comparisons.
a Significant compared to HC
b Significant compared to MCI
Table 2: Hippocampal subfield differences in AD, MCI, and HC subjects.
The highest diagnostic efficacy for discriminating AD subjects from HC or MCI subjects were right subiculum, right presubiculum, right molecular layer, right fimbria, and right whole hippocampus volumes. The similar pattern of subfield atrophy was also displayed in the left molecular layer, left fimbria, left whole hippocampus volumes in AD subjects relative to HC or MCI subjects. Only left fimbria volume was significantly reduced in MCI subjects compared to HC (normalized volume 47.7, sensitivity=66.7%, specificity=89.7%, AUC 0.709). The diagnostic performance of each subfield volume was shown in (Table 3 and Table 4).
| Normalized volume | Sensitivity (%) | Specificity (%) | AUC | LR+ | LR- | |
|---|---|---|---|---|---|---|
| Lt. Hippocampal_tail | 290.1 | 73.3 (44.9-92.2) | 96.6(82.2-99.9) | 0.926 | 21.3 | 0.3 |
| Rt. Hippocampal tail | 312.4 | 73.3 (44.9-92.2) | 93.1(77.2-99.2) | 0.871 | 10.6 | 0.3 |
| Lt. subiculum | 244.8 | 86.7 (59.5-98.3) | 96.6(82.2-99.9) | 0.959 | 25.1 | 0.1 |
| Rt. subiculum | 262.3 | 86.7 (59.5-98.3) | 90.0 (72.6-97.8) | 0.940 | 8.4 | 0.2 |
| Lt. CA1 | 370.8 | 80.2 (51.9-95.7) | 82.8 (64.2-94.2) | 0.890 | 4.6 | 0.2 |
| Rt. CA1 | 403.1 | 86.7 (59.5-98.3) | 82.8 (64.2-94.2) | 0.885 | 5.0 | 0.2 |
| Lt. presubiculum | 170.5 | 86.7 (59.5-98.3) | 96.6 (82.2-99.9) | 0.931 | 25.1 | 0.1 |
| Rt. presubiculum | 166.2 | 93.3 (68.1-99.8) | 93.1 (77.2-99.2) | 0.954 | 13.5 | 0.1 |
| Lt. parasubiculum | 28.6 | 80.0 (51.9-95.7) | 79.3 (60.3-92.0) | 0.782 | 3.9 | 0.3 |
| Rt. parasubiculum | 28.6 | 66.7 (38.4-88.2) | 96.6 (82.2-99.9) | 0.839 | 19.3 | 0.4 |
| Lt. molecular_layer_HP | 320.1 | 86.7 (59.5-98.3) | 96.6 (82.2-99.9) | 0.977 | 25.1 | 0.1 |
| Rt. molecular_layer_HP | 327.1 | 86.7 (59.5-98.3) | 96.6 (82.2-99.9) | 0.938 | 25.1 | 0.1 |
| Lt. GC-ML-DG | 166.6 | 93.3 (68.1-99.8) | 93.1 (77.2-99.2) | 0.945 | 13.5 | 0.1 |
| Rt. GC-ML-DG | 176.1 | 86.7 (59.5-98.3) | 96.6 (82.2-99.9) | 0.910 | 25.1 | 0.1 |
| Lt. CA3 | 109.2 | 66.7 (38.4-88.2) | 90.0 (72.6-97.8) | 0.844 | 6.4 | 0.4 |
| Rt. CA3 | 124.5 | 80.0 (51.9-95.7) | 86.2 (68.3-96.1) | 0.841 | 5.8 | 0.2 |
| Lt. CA4 | 144.9 | 93.3 (68.1-99.8) | 93.1 (77.2-99.2) | 0.940 | 13.5 | 0.1 |
| Rt. CA4 | 145.9 | 80.0 (51.9-95.7) | 96.6 (82.2-99.9) | 0.894 | 23.2 | 0.2 |
| Lt. fimbria | 44.0 | 100.0 (78.2-100.0) | 93.1 (77.2-99.2) | 0.977 | 14.5 | 0.0 |
| Rt. fimbria | 40.2 | 80.0 (51.9-95.7) | 93.1 (77.2-99.2) | 0.933 | 11.6 | 0.2 |
| Lt. HATA | 34.7 | 86.7 (59.5-98.3) | 79.3 (60.3-92.0) | 0.846 | 4.2 | 0.2 |
| Rt. HATA | 39.5 | 86.7 (59.5-98.3) | 82.8 (64.2-94.2) | 0.874 | 5.0 | 0.2 |
| Lt. Whole_hippocampus | 1955.2 | 86.7 (59.5-98.3) | 96.6 (82.2-99.9) | 0.970 | 25.1 | 0.1 |
| Rt. Whole_hippocampus | 1951.5 | 86.7 (59.5-98.3) | 96.6 (82.2-99.9) | 0.931 | 25.1 | 0.1 |
Table 3: The diagnostic performance, comparison between AD and HC.
| Normalized volume | Sensitivity (%) | Specificity (%) | AUC | LR+ | LR- | |
|---|---|---|---|---|---|---|
| Lt. subiculum | 240.1 | 80.0 (51.9-95.7) | 92.3 (74.9-99.1) | 0.882 | 10.4 | 0.2 |
| Rt. subiculum | 242.9 | 73.3 (44.9-92.2) | 96.2 (80.4-99.9) | 0.895 | 19.1 | 0.3 |
| Lt. CA1 | 377.7 | 86.7 (59.5-98.3) | 73.1 (52.2-88.4) | 0.856 | 3.2 | 0.2 |
| Lt. presubiculum | 163.7 | 80.0 (51.9-95.7) | 96.2 (80.4-99.9) | 0.897 | 20.8 | 0.2 |
| Rt. presubiculum | 166.2 | 93.3 (68.1-99.8) | 92.3 (74.9-99.1) | 0.936 | 12.1 | 0.1 |
| Lt. molecular_layer_HP | 308.2 | 80.0 (51.9-95.7) | 92.3 (74.9-99.1) | 0.928 | 10.4 | 0.2 |
| Rt. molecular_layer_HP | 315.9 | 80.0 (51.9-95.7) | 92.3 (74.9-99.1) | 0.910 | 10.4 | 0.2 |
| Lt. GC-ML-DG | 166.6 | 93.3 (68.1-99.8) | 76.9 (56.4- 91.0) | 0.890 | 4.0 | 0.1 |
| Rt. GC-ML-DG | 169.3 | 80.0 (51.9-95.7) | 88.5 (69.8-97.6) | 0.867 | 6.9 | 0.2 |
| Lt. CA3 | 109.2 | 66.7 (38.4-88.2) | 88.9 (51.8-99.7) | 0.770 | 6 | 0.4 |
| Rt. CA3 | 124.5 | 80.0 (51.9-95.7) | 80.8 (60.6-93.4) | 0.823 | 4.2 | 0.3 |
| Lt. CA4 | 144.9 | 93.3 (68.1-99.8) | 84.6 (65.1-95.6) | 0.900 | 6.1 | 0.1 |
| Rt. CA4 | 145.9 | 80.0 (51.9-95.7) | 88.5 (69.8-97.6) | 0.859 | 6.9 | 0.3 |
| Lt. fimbria | 38.4 | 93.3 (68.1-99.8) | 84.6 (65.1-95.6) | 0.921 | 6.1 | 0.1 |
| Rt. fimbria | 32.2 | 66.7 (38.4-88.2) | 96.2 (80.4-99.9) | 0.877 | 17.3 | 0.4 |
| Lt. Whole_hippocampus | 1955.2 | 86.7 (59.5-98.3) | 88.5 (69.8-97.6) | 0.923 | 7.5 | 0.2 |
| Rt. Whole_hippocampus | 1924.3 | 80.0 (51.9-95.7) | 96.2 (80.4-99.9) | 0.908 | 20.8 | 0.2 |
Table 4: The diagnostic performance, comparison between AD and MCI.
Comparison between AD and HC
Right subiculum, right presubiculum, right molecular layer, and right fimbria volumes were better predictors than right whole hippocampal volume (Table 3).
Comparison between AD and MCI
Left molecular layer (normalized volume 308.2, sensitivity=80.0%, specificity=92.3%, AUC 0.928) volumes was better predictor than left whole hippocampal volume (normalized volume 1955.2, sensitivity=86.7%, specificity=88.5%, AUC 0.923) (Table 4).
Comparison between MCI and HC
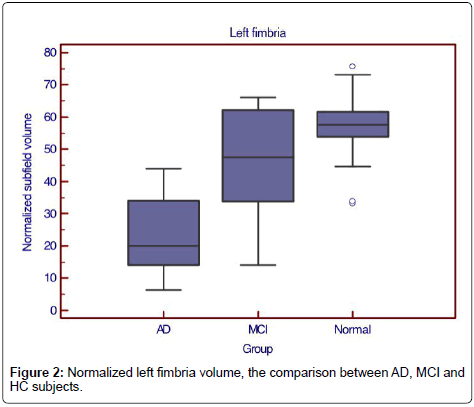
Only left fimbria volume was significantly reduced in MCI subjects compared to normal subjects (normalized volume 47.7, sensitivity=66.7%, specificity=89.7%, AUC 0.709, LR+6.4, LR- 0.4) (Figure 2).
When considered MCI with a positive amyloid PET, there were 5 of 8 cases defined as positive MCI. When using 48 as the cutoff value of the normalized volume of left fimbria to separate amyloid-PET positive MCI from HC, there were 4 cases correctly defined as positive MCI and 2 cases as negative MCI. The accuracy, false positive, and false negative of left fimbria as the predictor of PET-positive MCI were 75%, 12.5% and 12.5% respectively.
Discussion
The importance of a biomarker in clinical diagnosis for AD is not only the sensitivity but also the specificity. AD process is known to begin in limbic lobe structures from the deposition of neurofibrillary tangles causing neuronal loss and atrophy. Hippocampal volumetry is one of a biomarker for clinical diagnosis with high specificity. Furthermore, hippocampal subfield analysis could improve the diagnostic accuracy of AD based on neuropathological data which showing a differential vulnerability of hippocampal subfields to AD process.
Automated image analysis pipeline by Freesurfer version 6.0 is a combination of ex vivo and in vivo MRI data which much higher resolution than the in vivo atlas (v5.3) in exploring the subfields of the hippocampus, accurately tracing the molecular layer with hard dependence on geometric criteria and better-matched values to previously reported histological studies [20,21]. In this version, subfields CA2 and CA3 were combined to CA2/3 due to lack of distinguishing contrast on MRI. The limitation of FreeSurfer 5.3 subfield segmentation is due to being in vivo atlas’ delineation protocol, solely set up for the hippocampal body. It was not well interpreting the hippocampal head or tail and boundaries of the parcellation scheme which strongly differed from the majority of in vivo and imaging atlases. For example, FreeSurfer 5.3’s CA1 is the smallest subfield while CA2–3 is the biggest, compared with histologic data which shows the opposite.
Using automated analysis by Freesurfer version 6.0, we found that AD displayed a widespread pattern of subfield atrophy and right subiculum, right presubiculum, molecular layer, fimbria had high diagnostic accuracy. Significant subiculum, presubiculum and fimbria volume loss have been reported in previous studies using an early version of Freesurfer [22-24].However, the significant molecular layer volume loss in AD using Freesurfer version 6.0 in this study did not show in many previous studies due to the unavailable model of the molecular layer in FreeSurfer (v5.3 or earlier). Nevertheless, significant molecular layer volume loss in MCI compared to HC has been reported in a recent study using Freesurfer version 6.0. [13].
Only left fimbria volume was significantly reduced in MCI subjects compared to normal subjects in this study, same as a recent study [13]. Our study showed that the accuracy of left fimbria as the predictor of MCI converter was 75% when compared with amyloid PET result.
The fimbria is a white matter structure extending from the alveus and finally forming the fornix, containing myelinated axons (mostly efferent). The fornix and fimbria are sometimes referred to as the fimbria-fornix to reflect this functional unity and anatomic continuity. Callen et al. [25] found that almost all components of the limbic system, including the cross-sectional area of the fornix, showed a reduction in volume in patients with AD in comparison with age-matched controls. Copenhaver et al. [26] suggested that atrophy of the fornix and mammillary bodies might accompany the change from MCI to the development of clinical AD. Ringman et al. [27] followed this with diffusion tensor assessment of the fornix and orbitofrontal white matter projections. They found a significant reduction in Fractional Anisotropy (FA) in the fornix of patients with genetically inherited dementias in comparison with that in normal controls. The change of FA preceded the development of volume loss which can encourage early treatment as a preclinical biomarker identification.
Increased sensitivity of subfield volumetry more than total hippocampal volumetry to detect morphological changes at the MCI stage is consistent with NFT pathology, volumetric and neuronal loss. These are focal in the first pathophysiological stages and progressively extending to the whole hippocampus to become global in the advanced stage of dementia [28].
The cause of volume loss of the fimbria is not well understood. However, the recent study [13] assumed that it might be true biological process or the result of motion artifacts.
The demographic data showed the difference of TMSE between MCI and HC but not reached statistical significance. This may be due to small sample size and may explain about being no significant difference in other subfield volumes between these two groups.
In this study, Intracranial Volume (ICV) was used for normalized measured parameters because subjects with large ICV are expected to have larger hippocampi [29].The result of this study showed the better predictor for discriminating AD, MCI and HC groups in normalized group, for example, differentiating between AD and HC using normalized left fimbria volume (sensitivity=100.0%, specificity=93.1% and AUC=0.977) (Table 3) compared with left fimbria volume without ICV normalization (sensitivity=93.3%, specificity=96.6% and AUC=0.949).
The strengths of this study were a clinical evaluation by the senior geriatric neurologist (W.M) who specializes in dementia, complete neuropsychological test in memory aspect of each group and amyloid- PET result.
The study was limited by (1) small sample size, (2) using the atlas of only T1 data (using high-resolution T2 data giving good contrast between subregions, and segmentation of both channels simultaneously providing better information as demonstrated in the study of Iglesias et al. [13]), (3) no comparable MCI stable and MCI converter groups in this study which benefit for prediction MCI group who tended to develop AD in the future and might affect the treatment, (4) retrospective study design, and (5) some motion artifact in AD group. Further studies with larger sample size, a combination of T1/T2 data, and clinical follow up would be of interest to validate our findings in this study.
Conclusion
We demonstrated the possibility of using quantitative volumetry analysis of hippocampal subfield as a biomarker to differentiate AD from MCI and HC. This study was also demonstrating higher accuracy of some subfield volumes than the others. The left fimbria volume may be a good biomarker in early detection of MCI patients to be AD in the future.
Acknowledgment
The authors thank Mrs. Angkana Jongsawaddipatana and all staff members of the primary care unit of Siriraj Hospital for their contribution to data collection. The data of this study was partly from “the use of F-18 florbetapir (F-18-AV-45) PET to assess brain amyloid deposition in Alzheimer’s disease, mild cognitive impairment, and normal aging project” (code number Si137/2015) funded by Faculty of Medicine Siriraj Hospital, Mahidol University and National Research Council of Thailand. The authors (O.C, C.N, T.T, W.M) were funded by Chalermphakiat grant of Faculty of Medicine Siriraj Hospital.
References
- Alzheimer's Association (2014) 2014 Alzheimer's disease facts and figures. Alzheimers Dement 10: e47-92.
- Johnson KA, Fox NC, Sperling RA, Klunk WE (2012) Brain imaging in Alzheimer disease. Cold Spring Harb Perspect Med 2: a006213.
- Oishi K, Mielke MM, Albert M, Lyketsos CG, Mori S (2011) DTI analyses and clinical applications in Alzheimer's disease. J Alzheimers Dis 26 Suppl 3: 287-296.
- Johnson KA, Albert MS (2000) Perfusion abnormalities in prodromal AD. Neurobiol Aging 21: 289-292.
- Jack CR, Petersen RC, O'Brien PC, Tangalos EG (1992) MR-based hippocampal volumetry in the diagnosis of Alzheimer's disease. Neurology 42: 183-188.
- Fox NC, Warrington EK, Freeborough PA, Hartikainen P, Kennedy AM, et al. (1996) Presymptomatic hippocampal atrophy in Alzheimer's disease. A longitudinal MRI study. Brain 119: 2001-2007.
- Devanand DP, Pradhaban G, Liu X, Khandji A, De Santi S, et al. (2007) Hippocampal and entorhinal atrophy in mild cognitive impairment: prediction of Alzheimer disease. Neurology 68: 828-836.
- Apostolova LG, Dutton RA, Dinov ID, Hayashi KM, Toga AW, et al. (2006) Conversion of mild cognitive impairment to Alzheimer disease predicted by hippocampal atrophy maps. Arch Neurol 63: 693-699.
- Csernansky JG, Wang L, Swank J, Miller JP, Gado M, et al. (2005) Preclinical detection of Alzheimer's disease: hippocampal shape and volume predict dementia onset in the elderly. Neuroimage 25: 783-792.
- Frisoni GB, Sabattoli F, Lee AD, Dutton RA, Toga AW, et al. (2006) In vivo neuropathology of the hippocampal formation in AD: A radial mapping MR-based study. Neuroimage 32: 104-110.
- Ashburner J (2007) A fast diffeomorphic image registration algorithm. Neuroimage 38: 95-113.
- Wisse LE, Gerritsen L, Zwanenburg JJ, Kuijf HJ, Luijten PR, et al. (2012) Subfields of the hippocampal formation at 7 T MRI: in vivo volumetric assessment. Neuroimage 61: 1043-1049.
- Iglesias JE, Augustinack JC, Nguyen K, Player CM, Player A, et al. (2015) A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. Neuroimage 115: 117-137.
- Westman E, Aguilar C, Muehlboeck JS, Simmons A (2013) Regional magnetic resonance imaging measures for multivariate analysis in Alzheimer's disease and mild cognitive impairment. Brain Topogr 26: 9-23.
- Kälin AM, Park MT, Chakravarty MM, Lerch JP, Michels L, et al. (2017) Subcortical shape changes, hippocampal atrophy and cortical thinning in future Alzheimer's disease patients. Front Aging Neurosci 9: 38.
- Thientunyakit T, Sethanandha C, Muangpaisan W, Chawalparit O, Arunrungvichian K, et al. (2018) Implementation of [18F]-labeled amyloid brain PET imaging biomarker in the diagnosis of Alzheimer's disease: First-hand experience in Thailand. Nucl Med Commun 39: 186-192.
- http://adni.loni.usc.edu/wpcontent/uploads/2010/%2005/ADNIGO%20_PET_%20Tech%20_Manual_01142011.pdf
- http://adni.loni.usc.edu/Scientists/doc/ADNI2_PET%20Tech_Manualversion_4_2014Oct27_CLEAN.pdf
- Minoshima S, Drzezga AE, Barthel H, Bohnen N, Djekidel M, et al. (2016) SNMMI procedure standard/EANM practice guideline for amyloid PET imaging of the brain 1.0. J Nucl Med 57: 1316-1322.
- Simić G, Kostović I, Winblad B, Bogdanović N (1997) Volume and number of neurons of the human hippocampal formation in normal aging and Alzheimer's disease. J Comp Neurol 379: 482-494.
- Harding AJ, Halliday GM, Kril JJ (1998) Variation in hippocampal neuron number with age and brain volume. Cereb Cortex 8: 710-718.
- Khan W, Westman E, Jones N, Wahlund LO, Mecocci P, et al. (2015) Automated hippocampal subfield measures as predictors of conversion from mild cognitive impairment to Alzheimer's disease in two independent cohorts. Brain Topogr 28: 746-759.
- Lim HK, Hong SC, Jung WS, Ahn KJ, Won WY, et al. (2013) Automated segmentation of hippocampal subfields in drug-naïve patients with Alzheimer disease. AJNR Am J Neuroradiol 34: 747-751.
- Li YD, Dong HB, Xie GM, Zhang LJ (2013) Discriminative analysis of mild Alzheimer's disease and normal aging using volume of hippocampal subfields and hippocampal mean diffusivity: An in vivo magnetic resonance imaging study. Am J Alzheimers Dis Other Demen 28: 627-633.
- Callen DJ, Black SE, Gao F, Caldwell CB, Szalai JP (2001) Beyond the hippocampus: MRI volumetry confirms widespread limbic atrophy in AD. Neurology 57: 1669-1674.
- Copenhaver BR, Rabin LA, Saykin AJ, Roth RM, Wishart HA, et al. (2006) The fornix and mammillary bodies in older adults with Alzheimer's disease, mild cognitive impairment, and cognitive complaints: A volumetric MRI study. Psychiatry Res 147: 93-103.
- Ringman JM, O'Neill J, Geschwind D, Medina L, Apostolova LG, et al. (2007) Diffusion tensor imaging in preclinical and presymptomatic carriers of familial Alzheimer's disease mutations. Brain 130: 1767-1776.
- de Flores R, La Joie R, Chételat G (2015) Structural imaging of hippocampal subfields in healthy aging and Alzheimer's disease. Neuroscience 309: 29-50.
- Mueller SG, Schuff N, Yaffe K, Madison C, Miller B, et al. (2010) Hippocampal atrophy patterns in mild cognitive impairment and Alzheimer's disease. Hum Brain Mapp 31: 1339-1347.
Citation: Chawalparit O, Wontaneeporn N, Muangpaisan W, Thientunyakit T, Charnchaowanish P, et al. (2019) MRI Hippocampal Subfield Volume Analysis: Comparison between Alzheimer’s Disease, Mild Cognitive Impairment, and Normal Aging Subjects in an Amyloid PET Project. J Alzheimers Dis Parkinsonism 9:459. DOI: 10.4172/2161-0460.1000459
Copyright: © 2019 Chawalparit O, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 4983
- [From(publication date): 0-2019 - Sep 29, 2025]
- Breakdown by view type
- HTML page views: 4123
- PDF downloads: 860