Molecular Mechanism of High Glucose Induced Mitochondrial DNA Damage in Retinal Ganglion Cells
Received: 10-Aug-2021 / Accepted Date: 30-Aug-2021 / Published Date: 06-Sep-2021 DOI: 10.4172/1165-158X.1000203
Abstract
Background: Progressive death of retinal ganglion cells (RGCs) is the main pathological basis of glaucoma. It is found that mitochondrial DNA damage in RGCs may be closely related to lesion of glaucoma.
Methods: RGCs were cultured with different concentration of glucose and divided into 3 groups, namely NC group, Low-Glu group, High-Glu group. Cell counting kit-8 was used to detect the cell viability. Flow cytometry was used to detect the cell apoptosis. The DNA damage was measured with comet assay, and the morphological changes of damaged mitochondria in RGCs were observed by transmission electron microscopy (TEM). Western blot analyzed the expression of MRE11, RAD50, and NBS1 protein.
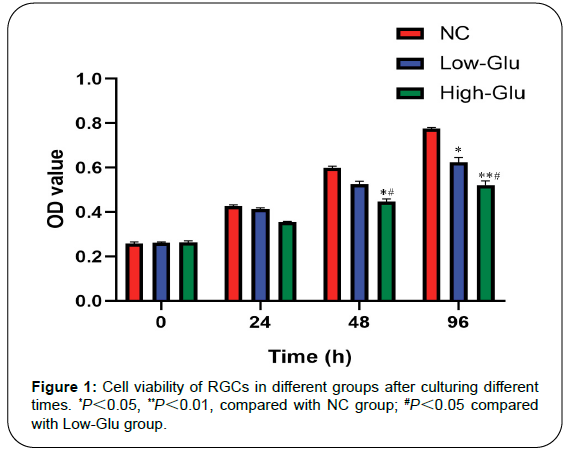
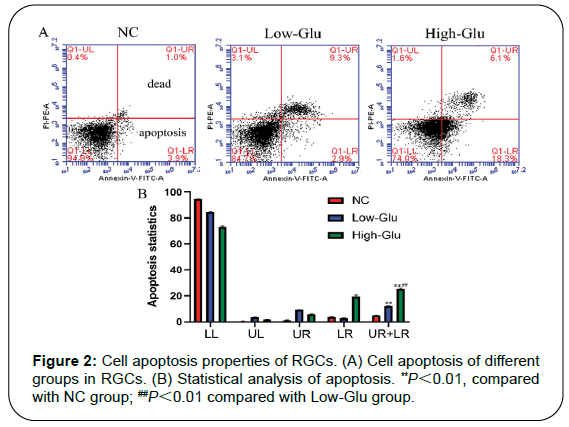
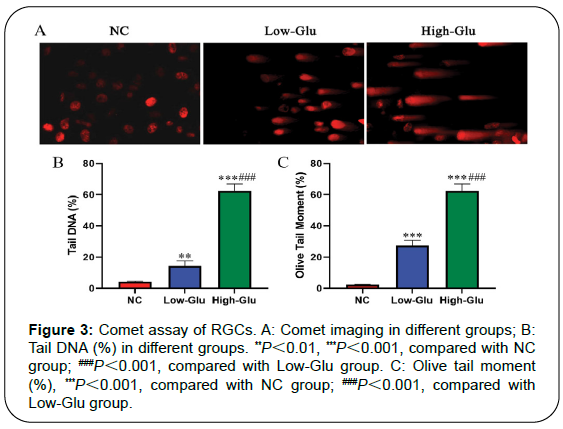
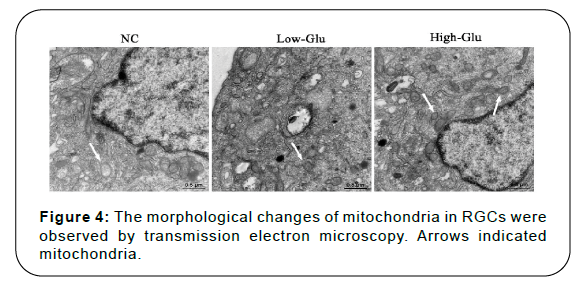
Results: Cell viability of RGCs in Low-Glu and High-Glu groups were lower than that in NC group in 48 and 96 h, *P<0.05, **P<0.01. The cell apoptosis in NC group was 4.9%, the Low-Glu group was 12.2% and High-Glu group was 24.4%. The comet imaging showed that NC cells did not have tailings, but the low-Glu and high-Glu group cells had tailings, indicating that the DNA of RGCs had been damaged. TEM showed that RGCs cultured with high glucose occurred mitochondrial morphology changes and dysfunction. Expression of MRE11, RAD50, NBS1 proteins associated with DNA damage repair pathway in High-Glu group were declined compared with Low-Glu group, #P<0.05.
Conclusion: High glucose induces mitochondrial DNA damage and results in apoptosis of retinal ganglion cells in glaucoma.
Keywords: Glaucoma; Retinal ganglion cells; High glucose; Mitochondria; DNA damage
Introduction
Glaucoma is one of the most common ocular diseases in the world. Its pathological manifestations are progressive retinal ganglion cells (RGCs) apoptosis and irreversible degeneration of the optic nerve. The onset of glaucoma is urgent, and the condition is serious. The patient will be easily blind if not be treated in time [1,2]. At present, the pathogenesis of glaucoma is not fully understood [3,4]. It has been reported than many patients with glaucoma will also have diabetes. The increased blood viscosity due to the elevation of blood glucose in diabetic patients affects the ocular vascular microcirculation and aqueous humor circulation, leading to increased intraocular pressure to induce glaucoma [5].
Diabetes can increase the apoptosis rate of retinal neurons. A large number of studies have confirmed that retinal neurons begin to undergo apoptosis before microvascular lesions occur. RGCs are the earliest retinal cells to be affected, and the apoptosis rate is significantly increased [6,7]. Functional tests in people with early diabetes can also detect the loss of retinal nerve cells. RGCs are nerve cells in the final segment of the retina and their dendrites are the first to be affected during the development of diabetes [8]. With the development of the disease, RGCs begin to undergo apoptosis, similar to central nerve cells, with a small amount of apoptosis in the early stage. The surrounding cells can fill the space by expanding their cell body and dendrites to compensate for their functions. However, with the increase of the number of cell apoptosis, a large number of blank areas will appear and the function will be decompensated [9]. RGCs are the only cells in the retina that can transmit visual signals to the visual center through axons [10]. With the progression of diabetes, increased apoptosis cells can accelerate the loss of RGCs, which will lead to the decline of visual acuity or even permanent loss [11,12].
Progressive death of RGCs is the main pathological basis of glaucoma [13]. Studies have found that mitochondrial dynamics damage may be closely related to RGCs damage of glaucoma [14,15]. Mitochondrial dynamics refers to the continuous division, fusion, movement, transportation and autophagy of mitochondria in cells, which have important effects on the survival, metabolism and maintenance of physiological functions of cells [16]. Although we currently have a preliminary understanding of the process of regulating mitochondrial dynamical damage in RGCs, the molecular mechanism of mitochondrial DNA damage in RGCs is still limited. In order to further understand the related molecular mechanisms of mitochondrial DNA damage in RGCs leading to glaucoma, the present study used hyperglycemic RGCs model induced by high glucose to investigate the related cellular molecular mechanisms.
Materials and Methods
Materials
Mouse retinal ganglion cells (RGCs) were purchased from Chinese Academy of Sciences; fetal calf serum (GIBCO); penicillin and streptomycin (100×) (Solarbio); 0.25% trypsin (Solarbio); Rat anti Human primary antibody MRE11, RAD50, NBS1 (1:500), HRP labeled Sheep anti Rat secondary antibody (1:5, 000) (Beyotime Biotechnology); CCK-8 (SAB); cell culture dish: 6/12/48 plate (Thermo fisher); sterile pipette: 5 mL/10 mL (Costar, USA); liquid transfer gun: 10 μL/20 μL/200 μL/1000 μL (Eppendorf); centrifuge tube: 10 mL/50 mL (Thermo fisher); centrifuge with low temperature and high speed (Eppendorf, USA); confocal microscope (Olyplus, Japan); leica optical microscope (Leica, German); enzyme label analyzer (Beijing Pulang New Technology Co., Ltd, China); western blot instruments (Eppendorf); flow cytometry (Thermo fisher).
Methods
Cell viability: RGCs were cultured with DMEM with 10% FBS and 1% penicillin and streptomycin (100×) in 5% CO2 incubator at 37°C. The cells in the logarithmic growth phase were digested by trypsin and counted under the microscope to prepare 1-5 ×104 cell/mL of cell suspension. Cell suspension (100 μL) were cultured in 96-well plates and divided into 3 group, namely normal control group (NC), low glucose group (Low-Glu) and high glucose group (High-Glu). Cell counting kit-8 (CCK-8) and serum-free essential basic medium were mixed according to 1:10 volume ratio, and 100 μL of mixture was added into the RGCs after culturing 0, 24, 48 and 96 h. The RGCs were continuously incubated in an incubator for 1 h at 37°C, and the absorbance at 450 nm was determined by enzyme plate analyzer. The optical density (OD) value of each test hole is subtracted from the OD value of the zeroing hole or the control hole. The OD value of each repeating hole is averaged.
Cell apoptosis: RGCs were cultured with DMEM contained 10% FBS, 1% penicillin and streptomycin (100×) in 5% CO2 incubator at 37°C. The cells in the logarithmic growth phase were digested by trypsin and divided into 3 group, namely NC group, Low-Glu group and High- Glu group. The cell culture medium of NC group was supplemented with the same volume of normal saline. The cell culture medium of Low-Glu group was supplemented with 15 mmol/L glucose solution. The cell culture medium of High-Glu group was supplemented with 35 mmol/L glucose solution. Each group of cells was cultured for 96 h. All cells were digested and collected, then supernatant was discarded after centrifuging at 1000 g for 5 min. 195 μL Annexin V-FITC solution was added to gently resuspend the cells. After gently mixing, 10 μL propidium iodide staining solution was added for culturing 10-20 min. Flow cytometry was used to detect cell apoptosis in different groups. Annexin V-FITC is green fluorescence and its corresponding detection channel is flow cytometer FL1. Propidium iodide (PI) is red fluorescence and its corresponding detection channel is flow cytometer FL2.
Comet assay: RGCs (2×105 cell/mL) were put in EP tube at 37°C water bath. The dissolved 1% normal melting point agarose (NMPA) was spread on the slide and condensed at 4°C for 10 min. The low melting point agarose (LMPA) was heated to dissolve the agarose completely and 1.0% concentration was prepared. 100 μL cell suspension was dissolved in 100 μL 1%LMPA, then dropped on the first layer of glue, and placed at 4°C to condense for 15 min to solidify the agarose. After the agarose cell gel solidified, the slides were slowly placed into the 4°C dyeing tank to crack for 20 h [17]. The slides were placed in an electrophoresis tank at room temperature for 20 min. Electrophoresis was performed at 4°C for 20 min with 25 V voltage and 300 mA. The slides were removed, and each slide was stained with 10 μg/mL propidium bromide (PI) for 20 min in a dark condition, and then decolorized with distilled water for 10 min. Green light (wavelength: 515-560 nm) was excited and photographed under a fluorescence microscope. The main detection indexes are tail DNA (%) and olive tail moment (OTM). Tail DNA% represents the percentage of tail DNA in total DNA content. OTM is the ratio of the difference between the tail optical density center and the head optical density center to tail DNA content.
Morphological changes of mitochondria: RGCs (2×105 cell/mL) were culture in 96-well plate with DMEM contained 10% FBS and 1% penicillin and streptomycin (100×), and divided into 3 groups. The cell culture medium of NC group was supplemented with the same volume of normal saline. The cell culture medium of Low-Glu group was supplemented with 15 mmol/L glucose solution. The cell culture medium of High-Glu group was supplemented with 35 mmol/L glucose solution. Each group of cells was cultured for 96 h. The ultrastructural and morphological changes of mitochondria in RGCs were observed by transmission electron microscopy.
Western blot analysis of MRE11, RAD50, NBS1 expression: RGCs were collected and added RIPA lysate (containing protease inhibitor) to fully crack on ice for 3-5 min. The lysate was transferred into a 1.5 mL centrifuge tube and centrifuged with 10000 r/min for 5 min at 4°C. Supernatant was absorbed and put into 1.5 mL centrifuge tubes, and the protein concentration was determined by BCA kit. Prepare 8% SDS-PAGE electrophoresis and the concentrated glue run for 60 min under 80 V. Separation glue run for 120 min under 120 V, then transferred membrane 150 min under 120 V, and sealed for 60 min. Primary antibodies rat anti human primary antibody MRE11, RAD50, NBS1 (1:500) were incubated with the RGCs at 4°C overnight. Second antibody HRP labeled sheep anti rat secondary antibody (1:5, 000) were incubated at room temperature for 1 h. Image J software analyzed the gray values of the strips.
Statistical analysis
SPSS19.0 statistical software was adopted for data statistical analysis. One-way analysis of variance (ANOVA) was used to express as the mean ± standard deviation (SD). Statistical significance was defined as P< 0.05.
Results
Cell viability
The activities of cells were showed by measuring the OD value of each group. CCK-8 contains WST-8, which is reduced by dehydrogenase in mitochondria to form yellow formazan with high water solubility. The number of formazan generated is proportional to the number of living cells, and the number of OD value of living cells is determined at 450 nm. The larger the OD value is, the stronger cell activity is. From 0 to 24 h, there was no statistically significant difference of cell viability of RGCs in NC, Low-Glu, and High-Glu group, P>0.05. However, in 48 h and 96 h, there was a significant difference among the three groups. Cell viability of RGCs in Low-Glu and High-Glu groups were lower than that in NC group, *P<0.05, **PFigure 1).
Cell apoptosis
There were 4 areas, and area in top right corner (UR) showed the late apoptotic cells, and area in lower right corner (LR) showed the early apoptotic cells. The sum of the percentages of apoptotic cells (UR+LR) indicated the total cell apoptotic ratio. The sum percentage of the late apoptotic cells and the early apoptotic cells in NC group was 4.9%, the Low-Glu was 12.2% and High-Glu was 24.4%. There was a significant difference between Low-Glu and High-Glu group, as shown in Figures 2A and 2B.
Comet assay detects DNA damage
Comet assay, also known as single cell gel electrophoresis (SCGE), is a simple and effective method to evaluate cell DNA damage (Figure 3). Typical comet imaging of each group is shown in Figure 4A. No tail was found in RGCs of NC group, but tail was found in Low-Glu and High-Glu group. It indicated that RGCs had different degrees of DNA damage, and the High-Glu group was the most serious. The comet experimental imaging was analyzed by CASP software. Tail DNA% and OTM were used as the index data of DNA damage, as shown in Table 1.
| Group | Tail DNA (%) | OTM (%) |
|---|---|---|
| NC | 4.27 ± 0.36 | 2.31 ± 0.13 |
| Low-Glu | 14.36 ± 1.85** | 27.49 ± 3.14** |
| High-Glu | 30.24 ± 2.74***## | 62.34 ± 4.47***## |
Table 1: Tail DNA and olive tail moment in comet assay of each group (Mean ± SD).
Morphological changes of mitochondria
Mitochondria in normal control group had uniform size, complete structure, orderly in arrangement, and good integrity of intercellular connections. However, after high glucose treatment, the mitochondria in RGCs were different in size and disordered in arrangement, and the cell structure had degenerative changes. The number of mitochondria was relatively reduced, and most of them had become vacuolated. The integrity of intercellular connections in High-Glu group was worse than that of NC and Low-Glu group, as shown in Figure 4. These results indicated that high glucose caused mitochondrial dysfunction and induced cell apoptosis.
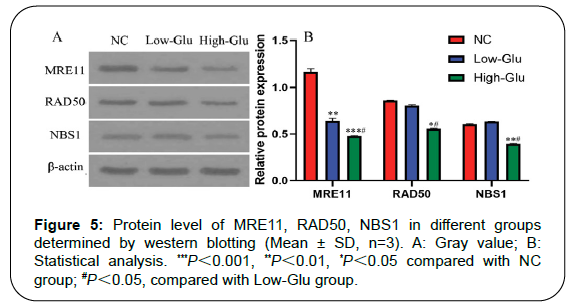
Expression of MRE11, RAD50, NBS1 protein
MRN is a complex discovered during the study of DNA double stranded damage repair pathway. It consists of three proteins, namely MRE11, RAD50 and NBS1. MRN complex plays an important role in the process of double-stranded DNA damage repair, participating in each link of DNA damage repair response such as initiation of repair, initiation of signal transduction and assistance in repair process. Western blot detected the protein expression of MRE11, RAD50 and NBS1 in each group, as shown in Figure 5. The protein content of MRE11, RAD50 and NBS1 decreased after RGCs incubating with glucose for 96 h, especially the High-Glu group. Compared with NC group, there was a significant difference in Low-Glu group and High- Glu group, ***P<0.001, **P<0.01, *P
Discussion
In the process of RGCs apoptosis, mitochondria, caspase protease family, signal transduction system, apoptosis-related genes, oxygen free radicals all participate in the occurrence of apoptosis. Mitochondria are also the source and object of oxidative stress and energy, so they play a decisive role in cell apoptosis. Mitochondria are the integrated elements of apoptosis pathway and the executors of cell death [18].
In this study, it was found that the structure of mitochondria in RGCs cultured with high glucose was incomplete, and the mitochondria were often vacuolated. In addition, the intercellular connection morphology was incomplete, among which the most serious was the cells in high-Glu group. These results indicate that high glucose induces morphological changes and abnormal function of mitochondria. Mitochondria are the main sites of intracellular oxidative phosphorylation and ATP formation, which provide the main energy for cells. Therefore, mitochondrial dysfunction is the key factor to induce cell apoptosis [19].
The special structure of mitochondrial DNA makes it vulnerable to attack. Double-stranded DNA damage is the main cause of genomic instability and cell death. MRE11/RAD50/NBS1 is an important complex for detection and repair of double-stranded DNA [20]. Replication, mitosis, and inheritance in cell metabolism can cause DNA damage after toxic chemicals and radiation therapy [21,22]. MRN complex plays an important role in the process of DNA damage repair, participating in each link of DNA damage repair response such as initiation of repair, initiation of signal transduction and assistance in repairment [23]. In recent years, it has been found that inhibiting the activity of MRN complex and hindering the repair of DNA damage in tumor cells can achieve the therapeutic effect of cancer [24]. In this research, western blot detected the lowest expression of MRE11/ RAD50/NBS1 in the High-Glu group. It is further proved that high glucose can damage the mitochondrial DNA structure of retinal ganglion cells in glaucoma, affect mitochondrial function and induce cell apoptosis. DNA is easy to be damaged by free radical because it is not protected by related proteins and is not easy to be repaired after damage. DNA oxidative damage and its mutations will hurt oxidative phosphorylation genes, and make enzymes related to the respiratory chain abnormal. This process results in mitochondrial oxidative phosphorylation dysfunction and promotes the production of reactive oxygen species, which further unbalances the respiratory chain and eventually leads to cell death [25].
The optic nerve damage in diabetic glaucoma is the result of multiple factors such as hyperglycemia and superoxidation damage. There is no evidence for retinal ganglion cells to regenerate. Retinal neuroprotective therapy is directly aimed at saving retinal ganglion cells. Therefore, to control blood glucose in diabetic patients and improve mitochondrial activity will become a new target for the treatment of glaucoma, which will also be a direction of future study by glaucoma researchers.
Conclusion
High glucose induces mitochondrial DNA damage in retinal ganglion cells and the morphological changes in mitochondria. The expression of important proteins MRE11/RAD50/NBS1 was decreased in DNA damage recovery. Therefore, DNA is easily damaged by free radicals, and is difficult to be repaired after damage, leading to retinal ganglion cell apoptosis.
Ethics Approval and Consent to Participate
The ethic approval was obtained from the Ethic Committee of The First People’s Hospital of Guiyang.
Consent to Publish
All of the authors have Consented to publish this research.
Availability of Data and Materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Competing Interests
All authors declare no conflict of interest.
Funding
None.
Authors’ Contributions
Each author has made an important scientific contribution to the study and has assisted with the drafting or revising of the manuscript.
Acknowledgements
We would like to acknowledge everyone for their helpful contributions on this paper.
References
- Zhang, Q. L., He, C., Li, R. Z., Ke, Y., Sun, Y. D., & Wang, J. Q.nmiR-708 and miR-335-3p Inhibit the Apoptosis of Retinal Ganglion CellsnThrough Suppressing Autophagy.nJ Mol Neurosci., 2021;71(2): 284-292.
- Yang, X. J., Zeng, Q., & Tezel, G. G.nRegulation of distinct caspase-8 functions in retinal ganglion cells and astroglianin experimental glaucoma.nNeurobiol Dis., 2021;150: 105258
- The AGIS Investigators.nThe Advanced Glaucoma Intervention Study (AGIS): The relationship betweenncontrol of intraocular pressure and visual field deterioration.nAm J Ophthalmol., 2000;130: 429-440.
- Feiner, L., & Piltz-Seymour, J. R.nCollaborative Initial Glaucoma Treatment Study. Collaborative Initial GlaucomanTreatment Study: a summary of results to date.nCurr Opin Ophthalmol., 2003;14(2): 106-111.
- Wang, Y.Y., Wang, C. G., Qi, S. N., Liu, Z. X., Su, G. F., & Zheng, X. J.nInteraction between XRCC 1 gene polymorphisms and diabetes onnsusceptibility to primary open-angle glaucoma.nExp Biol Med (Maywood)., 2019;244(7): 588-592.
- Rudraraju, M., & Somanath, P. R.nPharmacological Inhibition of Spermine Oxidase Reduces Neurodegenerationnand Improves Retinal Function in Diabetic Mice.nJ Clin Med., 2020;9(2): 340.
- Ng, D. S., Chiang, P. P., Tan, G., Cheung, C. G., Cheng, C. Y., Cheung, C. Y.,net al.nRetinal ganglion cell neuronal damage in diabetes and diabetic retinopathy.nClin Experiment Ophthalmol., 2016;44(4): 243-250.
- Salvi, L., Plateroti, P., Balducci, S., Bollanti, L., Conti, F. G., Vitale, M., et al.nAbnormalities of retinal ganglion cell complex at optical coherence tomographynin patients with type 2 diabetes: a sign of diabetic polyneuropathy, notnretinopathy.nJ Diabetes Complications., 2016;30(3): 469-476.
- Zhang, M. Y., Chen, L. F., Xu, F., Jiang, L. I., Yan, W. Y., Kunwar, B., et al.nInvolvement of Upregulated P53-Induced Death Domain Protein in RetinalnGanglion Cells Apoptosis After Optic Nerve Crush.nCurr Mol Med., 2019;20(1): 51-59.
- RodrÃguez-Muela, N., Germain, F., Mariño, G., Fitze, P. S., & Boya, P.nAutophagy promotes survival of retinal ganglion cells after optic nerve axotomynin mice.nCell Death Differ., 2012;19(1): 162-169.
- Garcia-Ayuso, D., Pierdomenico, J., Viadl-Sanz, M., & Villegas-Pérez, M. P.nRetinal Ganglion Cell Death as a Late Remodeling Effect of PhotoreceptornDegeneration.nInt J Mol Sci., 2019;20(18): 4649.
- Park, S. H., Park, J. W., Park, S. J., Kim, K. Y., Chung, J. W., Chun, M. H., etnal.nApoptotic death of photoreceptors in the streptozotocin-induced diabetic ratnretina.nDiabetologia., 2003;46(9): 1260-1268.
- Almasieh, M., Wilson, A. M., Morquette, B., Vargas, J. L., & Polo, A. D.nThe molecular basis of retinal ganglion cell death in glaucoma.nProg Retinal Eye Res., 2012;31(2): 152-181.
- Angert, M., Duong-Polk, K. X., Scott, R. T., Kim, J. J., Kukhmazov, I., Ellisman,nM. H., et al. Intraocular pressure elevation induces mitochondrial fission andntriggers OPA1 release in glaucomatous optic nerve.nInvest Ophthalmol Vis Sci., 2008;49(11): 4903-4911.
- Knott, A. B., Perkins, G., Schwarzenbacher, R., & Bossy-Wetzel, E.nMitochondrial fragmentation in neurodegeneration.nNat Rev Neurosci., 2008;9(7): 505-518.
- Hu, X. X., & Dai, Y.nThe role of mitochondrial dynamics in retinal ganglion cells.nChin J Exp Ophthalmol., 2017;35(1): 74-78.
- Olive, P. L., & Banath, J. P.nThe comet assay: a method to measure DNA damage in individual cells.nNat Protoc., 2006;1(1): 23-29.
- Areiuch, V. G. A., Elquero, M. E., Poderoso, J. J., & Carreras, M. C.nMitochondrial regulation of cell cycle and proliferation.nAntioxid Redox Signal., 2012;16(10): 1150-1180.
- Zirpoli, H., Sosunov, S. A., Niatsetskaya, Z. V., Mayurasakorn, K., Kollareth, D.nJ. M., Serhanc, C. N., et al.nNPD1 rapidly targets mitochondria-mediated apoptosis after acute injectionnprotecting brain against ischemic injury.nExp Neurol., 2021;335: 113495.
- Wang, Q., Goldstein, M., Alexander, P., Wakeman, T. P., Sun, T., Feng, J. J., etnal.nRad17 recruits the MRE11-RAD50-NBS1 complex to regulate the cellularnresponse to DNA double-strand breaks.nEMBO J., 2014;33(8): 862-877.
- Michel, B., Ehrlich, S. D., & Uzest, M.nDNA double-strand breaks caused by replication arrest.nEMBO J., 1997;16(2): 430-438.
- Van den Bosch, M., Bree, R. T., & Lowndes, N. F.nThe MRNcomplex: coordinating and mediating the response to brokennchromosomes.nEMBO Rep., 2003;4(9): 844-849.
- D′Amours, D., & Jackson, S. P.nThe Mre11 complex: at the crossroads of DNA repair and checkpoint signalling.nNat Rev Mol Cell Biol., 2002;3(5): 317-327.
- Bala, M. A.nConcerted action of Nrf2-ARE pathway, MRN complex, HMGB1 andninflammatory cy-tokines-implication in modification of radiation damage.nRedox Biol., 2014;2: 832-846.
- Tanwar, M., Dada, T., Sihota, R., & Dada, R.nMitochondrial DNA analysis in primary congenital glaucoma.nMol Vis., 2010;16: 518-533.
Citation: Zhou J, Chen F, Yan A, Xia X (2021) Molecular Mechanism of High Glucose Induced Mitochondrial DNA Damage in Retinal Ganglion Cells. Cell Mol Biol 67: 203. DOI: 10.4172/1165-158X.1000203
Copyright: © 2021 Zhou J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 2107
- [From(publication date): 0-2021 - Apr 05, 2025]
- Breakdown by view type
- HTML page views: 1462
- PDF downloads: 645